All multicellular life begins as a single-celled egg, in other words, but a tiny, spherical bag of proteins. From this singular cell rises the world of others expected to construct a life form, with each new cell creating in the ideal place at the correct time to do an exact capacity in a joint effort with its neighbors.
This is a natural phenomenon that biologists want to understand completely. Now, scientists at the Harvard Medical School and Harvard University have found the findings that systematically revealed how one cell builds an entire organism. They profiled every cell in developing zebrafish and frog embryos.
Utilizing single-cell sequencing innovation, the examination groups traced the fates of individual cells over the initial 24 hours of the life of developing life. Their examinations uncover the far-reaching scene of which qualities are turned on or off and when as embryonic cells progress into new cell states and sorts.
Together, the discoveries speak to an index of hereditary recipes for creating diverse cells composed of two essential model species and give an extraordinary asset to investigating formative science and ailment.
Allon Klein, HMS assistant professor of systems biology, said, “With single-cell sequencing, we can, in a day’s work, recapitulate decades of painstaking research on the decisions cells make at the earliest stages of life.”
“Biomedically, these baseline resources for how organisms develop are as important as having baseline resources for their genomes.”
“With the approaches that we’ve developed, we’re charting what we think the future of developmental biology will be as it transforms into a quantitative, big-data-driven science.”
Scientists noted that their work could pave the way to a new understanding of a host of diseases.
Alexander Schier, the Leo Erikson Life Sciences Professor of Molecular and Cellular Biology at Harvard, said, “We foresee that any complex biological process in which cells change gene expression over time can be reconstructed using this approach. Not just the development of embryos but also the development of cancer or brain degeneration.”
During the study, scientists leveraged the power of InDrops, a single-cell sequencing technology. They captured quality gene-expression information from every cell of the developing life, each one in turn. The groups profiled in excess of 200,000 cells at various times, focusing more than 24 hours from the two species.
Courtesy of Enrique Amaya
To map the lineage of essentially every cell as an embryo creates, alongside the exact succession of quality articulation occasions that stamp new cell states and sorts, the groups grew new trial and computational systems, including the presentation of fake DNA scanner tags to track the genealogy connections between cells, called TracerSeq.
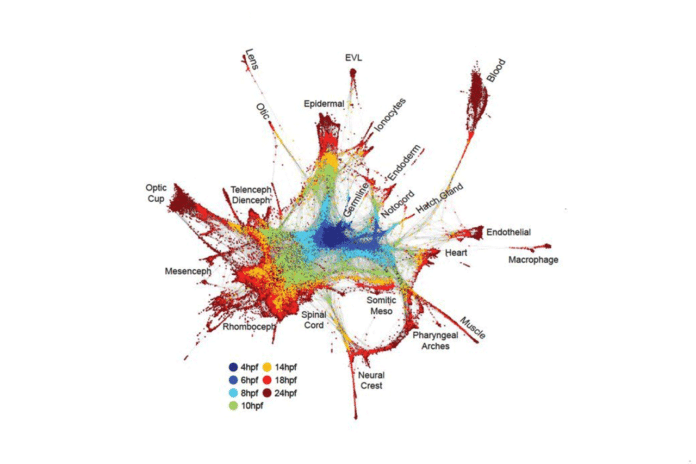
In the investigation co-drove by Schier, the exploration group utilized Drop-Seq — a solitary cell sequencing innovation created by analysts at HMS and the Broad Institute of MIT and Harvard — to consider zebrafish fetuses more than 12 hours at high time determination. Cooperating with Aviv Regev, a central part of the Broad, the group remade cell directions through a computational strategy they named URD, after the legendary Norse figure who chooses all fates.
Schier and associates profiled in excess of 38,000 cells and built up a cell “family tree” that uncovered how quality articulation in 25 cell writes changed as they concentrated. By joining that information with spatial surmising, the group was additionally ready to reproduce the spatial roots of the different cells written in the early zebrafish embryo.
In the two species, the groups’ discoveries reflected quite a bit of what was beforehand thought about the movement of embryonic advancement. This outcome underscored the energy of the new methodologies. Be that as it may, the investigations were extraordinary in uncovering in far-reaching subtle elements the falls of occasions that take cells from early forebear or “generalist” states to more particular states with barely characterized capacities.
The groups distinguished generally hard-to-recognize subtle elements, for example, uncommon cell composes and subtypes, and connected new and very particular quality articulation examples to various cell heredities. In a few cases, they discovered cell composes developing far sooner than beforehand had been thought.
For researchers endeavoring to answer inquiries regarding human disease, this information could be intensely enlightening. In regenerative drugs, for instance, specialists have, for a considerable length of time, intended to control immature microorganisms toward particular destinies with the objective of supplanting imperfect cells, tissues, or organs with practical ones. Recently gathered insights about the succession of quality articulation changes that hasten the rise of particular cell compose can move these endeavors further.
Klein said, “With these datasets, if someone wants to make a specific cell type, they now have the recipe for the steps that those cells took as they formed in the embryo. We’ve in some sense established a gold standard reference for how complex differentiation processes actually progress in embryos, and set an example for how to systematically reconstruct these types of processes.”
When combined with one of the core concepts in biological inquiry — the idea of disrupting a system to study what happens — single-cell sequencing can yield insights difficult to attain before, Klein said.
As a proof of principle, Klein, Megason, and colleagues used the CRISPR/Cas9 gene editing system to create zebrafish with a mutant form of chordin, a gene involved in determining the back-to-front orientation of a developing embryo. Schier and colleagues took a similar approach by profiling zebrafish with a mutation in a different patterning gene known as a one-eyed pinhead.
When analyzed with single-cell sequencing, the teams confirmed previously known descriptions of chordin and one-eyed pinhead mutants. They could describe in detail or even predict the effects of these mutations on developing cells and nascent tissues across the whole embryo.
Unexpectedly, the groups independently found that at the single-cell level, gene expression was the same in mutants and wild-type, despite the loss of an essential signaling pathway. The proportions of different cell types, however, changed.
Schier said, “This work only became possible through recent technologies that let us analyze gene expression in thousands of individual cells. Now the scale is much larger so that we can reconstruct the trajectory of almost all cells and all genes during embryogenesis. It is almost like going from seeing a few stars to seeing the entire universe.”
The examination groups additionally showed how this information could be mined to answer long-standing essential scientific inquiries.
Whenever Klein, Kirschner, Megason, and partners thought about cell-state scenes amongst zebrafish and frog incipient organisms, they generally watched similitudes. Be that as it may, their examinations uncovered various amazements also. One such perception was that qualities stamping cell states in a single animal group were frequently poor markers for a similar cell state in alternate species.
In a few occurrences, they found that the DNA succession of quality — and the structure of the protein it encoded — could be almost indistinguishable between species yet have altogether different articulation designs.
The reason these differences were not spotted before, the researchers hypothesize, is that computational analyses “pay attention” to data in a way fundamentally different from how humans do.
“I think this reflects some level of confirmation bias. When scientists find something conserved between species, they celebrate it as a marker,” Megason said. “But often, all the other nonconserved features are ignored. Quantitative data helps us move past some of these biases.”
In another striking finding, the groups watched that the procedure of cell separation into particular sorts — which is normally thought to happen in a tree-like structure where distinctive cell writes fan out from a typical precursor cell — can shape “circles” and, in addition, branches.
For instance, the neural peak — a gathering of cells that offer ascent to assorted tissue composed including smooth muscle, certain neurons, and craniofacial bone — at first rises up out of neural and skin forerunners; however, it is notable to produce cells that show up relatively indistinguishable to bone and ligament antecedents.
Klein said, “The new results suggest that similar loops might occur in other situations. That cells in the same state can have very different developmental histories suggests that our hierarchical view of development as a “tree” is far too simplified.”
Each of the three groups additionally recognized certain cell populaces that existed in a sort of halfway “basic leadership” state. Schier and partners found that at certain key formative branch focuses, cells seemed to go down one formative direction but then changed their destiny to another.
Klein, Megason, Kirschner, and partners mentioned a related objective fact that, right on time being developed, a few cells enacted two particular formative projects. In spite of the fact that those middle-of-the-road cells would inevitably embrace a solitary personality, these disclosures add to the photo of how cells build up their destiny and clue that there might be factors or past qualities associated with coordinating that.
Kirschner said, “With multilineage cells, we have to start wondering if their final fate is being determined by some selective force or interaction with the environment, rather than just genetic programs.”
The recently produced informational collections and the new instruments and advances created as a component of these examinations establish the framework for a wide range of future investigation, as indicated by the creators.
Formative scholars can accumulate progressively and higher-quality information on numerous species, take after incipient organisms assist in time, and play out any number of bother tries, all of which can help enhance comprehension of the principal guidelines of science and illness.
These assets can likewise fill in as a point of convergence for cooperation and association since most labs don’t have the profundity of mastery expected to abuse every one of the information and data created.
Kirschner said, “I think these studies are creating a real sense of community, with researchers raising questions and interacting with each other in a way that harkens back to earlier times in the study of embryology.”
Schier said, “The three studies are an example of how the scientific community can work on complementary questions to answer important questions in biology. Instead of competing, our groups were in regular contact over the past two years and coordinated the publication of our studies. And it is great how complementary the three papers are — each highlights different ways such complex data sets can be generated, analyzed and interpreted.”
Megason said, “Right now, we have a roadmap, but it doesn’t tell us what the signs are. What we need to do is figure out the signals that direct cells down certain roads, and what the internal mechanisms are that allow cells to make those decisions.”
Klein said, “The beauty of working on an organism is that this is it,” Klein said. “Ten, 20 years from now, we can still be sure zebrafish and frogs are going to develop according to the same patterns.”
Journal Reference
- James A. Briggs et al., The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018). DOI:10.1126/science.aar5780
- Jeffrey A. Farrell et al., Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131(2018). DOI:10.1126/science.aar3131
- Daniel E. Wagner et al., Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981-987 (2018). DOI:10.1126/science.aar4362