The covering of a cell, called the membrane, is not just a wall. It has many proteins that do essential jobs in the cell. To understand what these proteins do and how they work together, scientists need to know how they are arranged and how they talk to each other. However, finding this information takes much work.
Now, researchers from Yale University have made a new way to look at membranes. It’s called Native-nanoBleach. This method helps overcome the problems scientists face when understanding how proteins are organized in the membrane. It tackles issues like studying membranes without disturbing the natural setup and dealing with the limits of regular microscopes used for this research.
The researchers tested their new method on a long-standing problem related to proteins linked to pancreatic cancer development. They successfully used the technique to find answers that had puzzled scientists for many years. The details of this breakthrough are explained in a recent study published in Nature Nanotechnology.
The usual methods for studying how proteins are organized in cell membranes involve taking the proteins out of their natural environment and putting them into artificial settings that only partly resemble biological cell membranes. This removes important details because proteins naturally interact with the molecules around them.
Additionally, regular light microscopes, commonly used for observing protein organization, can’t show if nearby proteins interact or just close neighbors on the membranes because they lack the necessary precision.
Finally, the level of a specific protein in a cell membrane could sometimes be higher for current research methods. In such cases, scientists have to make changes. They might need to create more proteins that are too scarce naturally or separate proteins when there are too many. However, this process can lead to losing essential details about how proteins naturally exist and work in cell membranes.
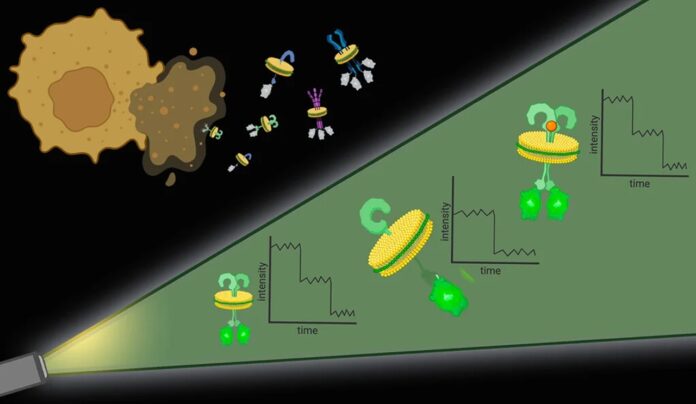
Bhattacharyya said, “Ideally, we would have a method that would work with any endogenous cell membrane protein expression level. To tackle the first challenge, the researchers used particular molecules and types of polymers to punch out protein complexes with their surrounding cell membrane intact. It’s like the cell membrane is a sheet of cookie dough, and the polymers are cookie cutters.”
This study introduces a new method to understand how membrane proteins, which comprise about 60% of drug targets, come together to function in their natural environment,” explained Gerard Walker, a graduate student in Bhattacharyya’s lab and co-first author of the paper.
To show how this method works, the researchers tackled a long-standing question in biology. They focused on a protein called KRas, often mutated in pancreatic cancers. The debate was whether KRas units form pairs (dimers) or groups (oligomers) on cell membranes. Previous studies, lacking molecular detail, suggested they come together, while biophysical analyses without the natural membrane indicated KRas remains as single units (monomers)
“Our method gives us the best of both worlds,” said Bhattacharyya. “We keep the natural membrane setting and achieve high precision in understanding the arrangement of molecules at a tiny scale. When we used our method, we discovered that KRas is present in similar amounts in both pairs (dimers) and single units (monomers). However, in pancreatic cancers where KRas is mutated, dimers increase, and monomers decrease.”
This discovery emphasizes the importance of studying proteins in their natural environment. It points to a potential target—reducing KRas pairs—for treating cancer. Bhattacharyya added that this method can be applied in various ways to understand how the organization of membrane proteins contributes to diseases.
“It’s great to see Native-nanoBleach working well for critical biological questions in our lab and beyond,” said Caroline Brown, a Ph.D. candidate in Kallol Gupta’s lab and co-first author of the study.
Bhattacharyya added, “A third of all human proteins are membrane proteins, and our method can be used to study any of them.”
“It’s a versatile technique,” she explained. “There are no restrictions.”
Looking ahead, Bhattacharyya and her team aim to use this method to investigate how proteins are organized in the membranes of different cell structures, such as mitochondria.
In conclusion, using molecular ‘cookie cutters’ in the form of Native-nanoBleach presents a breakthrough in studying membrane protein organization, offering a versatile and practical approach with potential applications in diverse biological investigations.
Journal reference:
- Walker, G., Brown, C., Ge, X. et al. Oligomeric organization of membrane proteins from native membranes at nanoscale spatial and single-molecule resolution. Nature Nanotechnology. DOI: 10.1038/s41565-023-01547-4.