Spatial transcriptomics and proteomics provide complementary information that transformed our understanding of complex biological processes. However, experimental Integration of these modalities is limited.
A new method called Spatial PrOtein and Transcriptome Sequencing (SPOTS) can illuminate the identities and activities of cells throughout an organ or a tumor at unprecedented resolution. Developed by researchers at Weill Cornell Medicine, NewYork-Presbyterian, and the New York Genome Center, SPOTS can illuminate the identities and activities of cells throughout an organ or a tumor at unprecedented resolution.
The technique preserves information about the precise locations of the cells while recording patterns of gene activity and the presence of essential proteins in cells across tissue samples. This makes it possible to create intricate, data-rich “maps” of organs, including organs that are ill and tumors, which may be extremely helpful in both basic and clinical research.
Study co-senior author Dr. Dan Landau, an associate professor of medicine in the Division of Hematology and Medical Oncology and a member of the Sandra and Edward Meyer Cancer Center at Weill Cornell Medicine and a core faculty member at the New York Genome Center, said, “This technology is exciting because it allows us to map the spatial organization of tissues, including cell types, cell activities, and cell-to-cell interactions, as never before.”
The new approach is a part of a larger initiative by scientists and engineers to create more effective means of “seeing” at the microscopic level how organs and tissues function. Recent years have seen significant advancements in research, notably in methods for profiling gene activity and other layers of data in single cells or small groups of cells. The information regarding the original placements of the profiled cells within the tissues needs to be recovered, however, because these approaches frequently ask for the breakdown of tissues and the separation of cells from their neighbors. The new technique also records that spatial data, and it does so with excellent resolution.
The method is based in part on existing 10x Genomics technology. It uses glass slides suitable for imaging tissue samples with ordinary microscope-based pathology methods but is also coated with thousands of particular probe molecules.
Each probe molecule’s chemical “barcode” identifies its two-dimensional location on the slide. The probe molecules on the slide grab the messenger RNAs (mRNAs), essentially the transcripts of activated genes, from neighboring cells when a thinly sliced tissue sample is placed on the slide, its cells are rendered permeable. Designer antibodies are used in the procedure, and they attach to the unique probe molecules and proteins of interest in the tissue.
Researchers can quickly and automatically identify the collected mRNAs and chosen proteins and precisely map them to their original locations throughout the tissue sample. The produced maps may be considered independently or in comparison to the sample’s routine pathology imaging.
On tissue from a healthy mouse spleen, the team used SPOTS to show the intricate functional architecture of this organ, including clusters of various cell types, their functional states, and how those states altered depending on the cells’ placements.
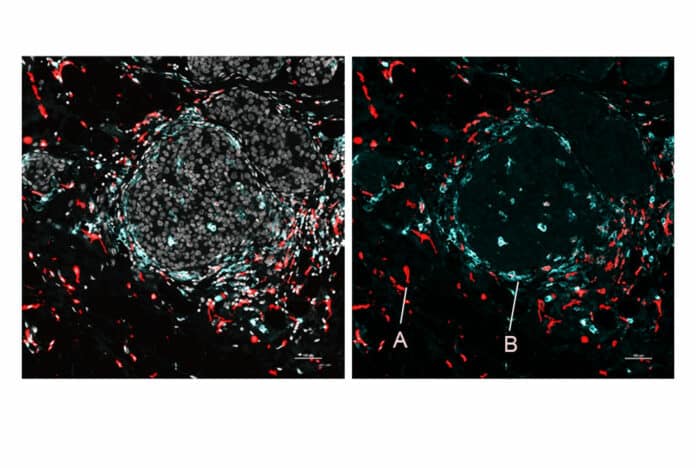
The researchers also utilized SPOTS to map the cellular structure of a mouse breast tumor, highlighting its potential for use in cancer research. The generated map showed macrophages, and immune cells, in two different states, each indicated by a different protein marker: one condition was active and battling tumors, while the other was immuno-suppressive and establishing a barrier to shield the tumor.
Dr. Landau, an oncologist at NewYork-Presbyterian/Weill Cornell Medical Center, said, “We could see that these two macrophage subsets are found in different areas of the tumor and interact with different cells—and that difference in the microenvironment is likely driving their distinct activity states.”
“Such details of the tumor immune environment—details that often can’t be resolved due to immune cells’ sparseness within tumors—might help explain why some patients respond to immune-boosting therapy and some don’t, and thus could inform the design of future immunotherapies.”
“This initial version of SPOTS has a spatial resolution such that each “pixel” of the resulting dataset sums gene activity information for at least several cells. However, the researchers hope soon to narrow this resolution to single cells while adding other layers of key cellular information.”
Journal Reference:
- Ben-Chetrit, N., Niu, X., Swett, A.D. et al. Integration of whole transcriptome spatial profiling with protein markers. Nat Biotechnol (2023). DOI: 10.1038/s41587-022-01536-3