Nanopore sequencing is a one-of-a-kind, adaptable innovation that empowers immediate, real-time analysis of long DNA or RNA fragments. It works by checking changes to electrical current as nucleic acids are go through a protein nanopore. The following sign is decoded to give the particular DNA or RNA sequence.
Scientists have continually tried to extend this principle and build larger pores to accommodate proteins for sensing purposes. Still, a significant challenge has been the limited understanding of artificial protein design. As an alternative, the DNA origami technique was invented.
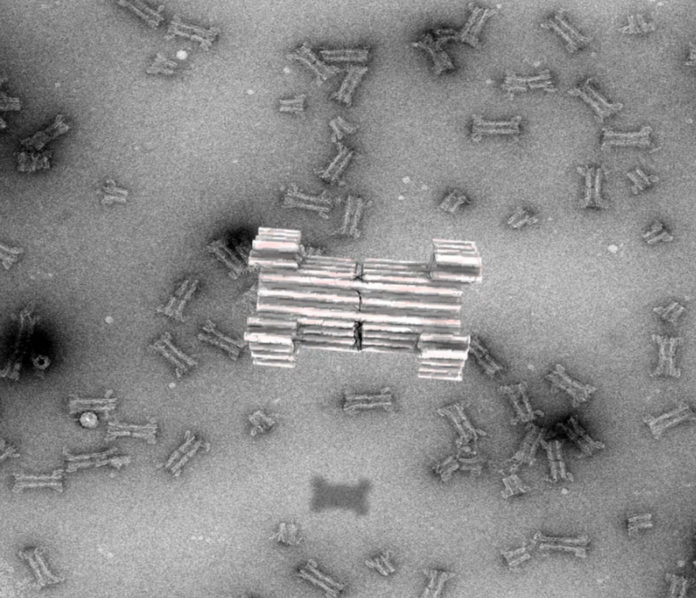
DNA Origami is one of the most recent techniques of utilizing DNA as a building block for the synthesis of nanoparticles.
Now, scientists from Aarhus University have reported the creation of a large synthetic nanopore made from DNA. This new synthetic nanopore can translocate large protein-sized macromolecules between compartments separated by a lipid bilayer.
It involves a functional gating system that enables the biosensing of a few molecules in solution. Moreover, a controllable plug in the pore made it possible to size-selectively control the flow of protein-size molecules and demonstrate label-free, real-time bio-sensing of a trigger molecule.
In addition to that, the pore is composed of a set of controllable flaps that allow targeted insertion into membranes displaying particular signal molecules.
According to scientists, this synthetic nanopore will enable the insertion of the sensor specifically into diseased cells and may allow diagnosis at the single-cell level.
Journal Reference
- Thomsen, R.P., Malle, M.G., Okholm, A.H. et al. A large size-selective DNA nanopore with sensing applications. Nature Communications 10, 5655 (2019). DOI: 10.1038/s41467-019-13284-1