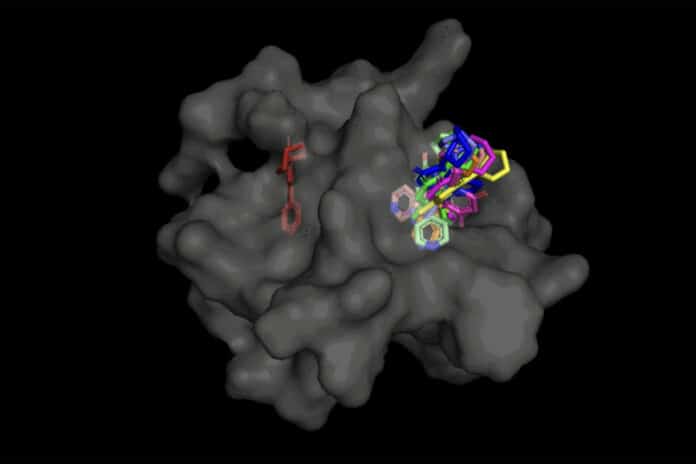
Molecular docking- a process for predicting the binding structure of a small molecule ligand to a protein- is essential for drug design. Recent deep-learning methods that treat docking as a regression problem have decreased runtime compared to traditional search-based methods but have yet to offer substantial improvements in accuracy.
There could be more to generative diffusion models, believes MIT scientists.
A team of researchers at MIT’s Abdul Latif Jameel Clinic for Machine Learning in Health (Jameel Clinic) have developed a new molecular docking model called DiffDoc. Given that most pharmaceutical companies now use state-of-the-art tools, the model’s distinctive approach to computational drug design represents a paradigm change and offers significant potential to restructure the current drug development process.
In the instance of DiffDock, the model can effectively detect various binding sites on proteins that it has never encountered after being trained on multiple ligand and protein configurations. The process creates fresh 3D coordinates rather than new picture data to assist the ligand in determining potential orientations for fitting into the protein pocket.
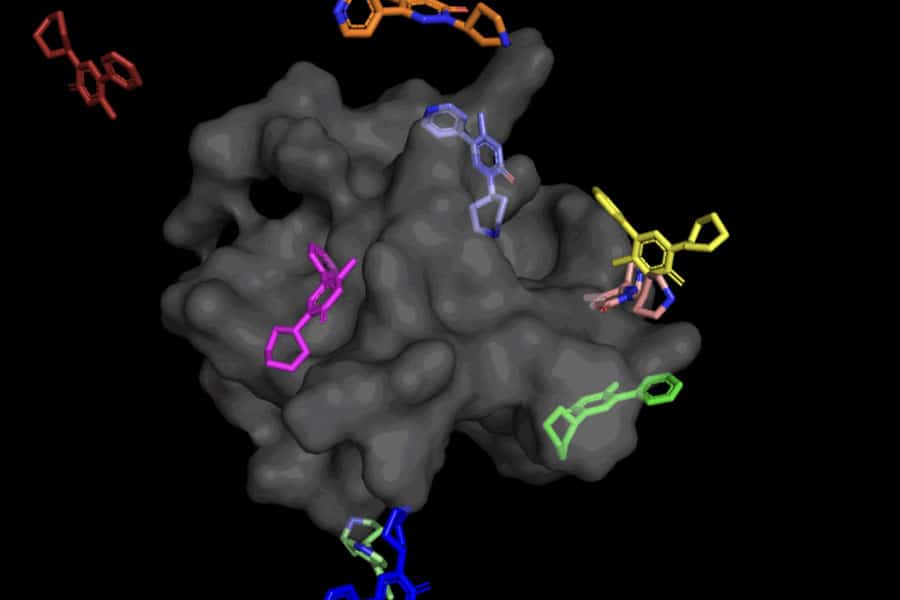
This “blind docking” method opens up new ways to use AlphaFold 2 (2020), DeepMind’s well-known protein folding AI model. The potential of AlphaFold’s computationally folded protein structures to assist in discovering novel pharmacological mechanisms of action has generated a significant deal of interest in the research community since the initial publication of AlphaFold 1 in 2018. Modern molecular docking methods, however, have yet to show that they are any more effective than chance at binding ligands to computationally predicted structures.
Because of its capacity to reason at a larger scale and implicitly incorporate part of the protein flexibility, DiffDock is not only much more accurate than past approaches to classic docking benchmarks, but it also continues to perform well even after other docking models start to falter. DiffDock places 2 angstroms (generally regarded as the threshold for an accurate pose, where one corresponds to one over 10 billion meters) or less in 22 percent of its predictions in the more realistic scenario involving the use of computationally generated unbound protein structures, more than double other docking models, which only reach 10 percent for some and fall as low as 1.7 percent.
These developments open up a whole new world of possibilities for biological study and medication development. For instance, phenotypic screening, which involves observing a drug’s effects on a disease without knowing which proteins it is working upon, is a common method for discovering new medications.
The drug’s mechanism of action must be identified to understand better its potential adverse effects and how it might be enhanced. This process, called “reverse screening,” can be very difficult and expensive. Still, a combination of protein folding techniques and DiffDock may allow performing a significant portion of the process in silico, allowing potential “off-target” side effects to be identified early on before clinical trials.
Tim Peterson, an assistant professor at the University of Washington St. Louis School of Medicine, says, “DiffDock makes drug target identification much more possible. Before, one had to do laborious and costly experiments (months to years) with each protein to define the drug docking. But now, one can screen many proteins and do the triaging virtually in a day.”
“There is a very ‘fate loves irony’ aspect that Eroom’s law — that drug discovery takes longer and costs more money each year — is being solved by its namesake Moore’s law — that computers get faster and cheaper each year — using tools such as DiffDock.”
Journal Reference:
- Gabriele Corso, Hannes Stark et al. DiffDock: Diffusion Steps, Twists, and Turns for Molecular Docking. DOI: 10.48550/arXiv.2210.01776