Copy number variants (CNVs) are a noteworthy reason for a few genetic disorders, making their identification an essential segment of genetic analysis pipelines. Current strategies for distinguishing CNVs from exome-sequencing data are restricted by high false-positive rates and low concordance due to inherent biases of individual algorithms.
To overcome these issues, calls generated by two or more algorithms are often intersected using Venn diagram approaches to identify “high-confidence” CNVs.
In a new study, scientists have presented a new machine-learning method to precisely identify deletions and duplications in the genome- called copy number variants- that mostly linked to autism and other neurodevelopmental disorders.
The method called CN-Learn integrates data from a few algorithms that endeavor to recognize copy number variations from exome-sequencing data— high-throughput DNA sequencing of just the protein-coding regions of the human genome.
Santhosh Girirajan, associate professor of biochemistry and molecular biology at Penn State and the lead author of the paper said, “Exome sequencing is fast becoming the gold standard for identifying genetic variations in clinical settings because it is faster and less expensive than other methods. However, current algorithms for identifying copy number variation from exome sequencing data suffer from very high false-positive rates — many of the variants they identify aren’t real. With our new method, called ‘CN-Learn,’ around 90% of the copy number variants we report are real.”
To recognize copy number variants from exome-sequencing data, specialists take a gander at the relative amount of DNA sequences delivered from each gene. If there is just one copy of a gene present in an individual, they hope to see less sequencing reads than if there are two copies, and three copies of a gene would prompt more reads.
In any case, it’s not exactly that simple, because various factors can impact what number of sequencing peruses are delivered from each gene. Specialists have, in this way built up a few algorithms to effectively recognize copy number variants from exome-sequencing data. Exclusively, in any case, these algorithms are not exceptionally reliable.
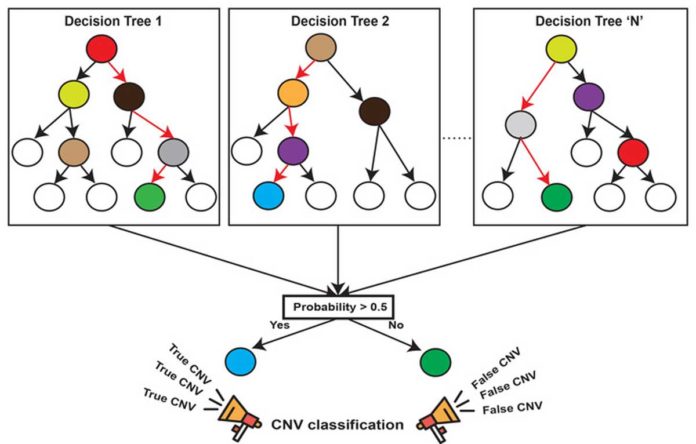
CN-Learn integrates data from four different copy-number-variant algorithms and uses a small set of biologically validated deletions and duplications to learn the signatures of these genomic events.
This learning process is facilitated by a machine-learning algorithm called ‘random forest,’ which uses hundreds of decision trees to model the relationship between the genetic context of deletions and duplications and the likelihood they are validated. CN-Learn then uses this model to predict deletions and duplications in other samples without validations.
Vijay Kumar Pounraja, a graduate student at Penn State and first author of the paper said, “Generally, the high number of false positives from copy-number-variant algorithms has been dealt with by using multiple algorithms and only counting the variants identified by all the methods — like a Venn diagram. This approach has multiple drawbacks and limitations, so we decided to develop a new machine-learning method instead.”
Santhosh Girirajan, associate professor of biochemistry and molecular biology at Penn State said, “Decisions about a patient’s diagnosis and eventual treatment are made based on this information, so it’s incredibly important to get them right. Because of this, we’ve made CN-Learn and all of the necessary supporting programs available to download in one easy package.”
The study was published in the journal Genome Research.