The advent of CRISPR-Cas9 revolutionized the field of genome editing, offering unprecedented precision and efficiency in manipulating genetic material. However, the application of this powerful tool in humans has encountered challenges due to potential off-target effects and limitations in delivery methods. Recent research has unveiled a new CRISPR-like system within animals that holds great promise for precise editing of the human genome. This groundbreaking discovery opens up a new avenue for exploring genetic modifications in a more controlled and targeted manner.
In this study, we delve into the characteristics and potential applications of this newly identified animal CRISPR-like system, shedding light on its implications for advancing gene editing technologies and understanding the intricate workings of the human genome.
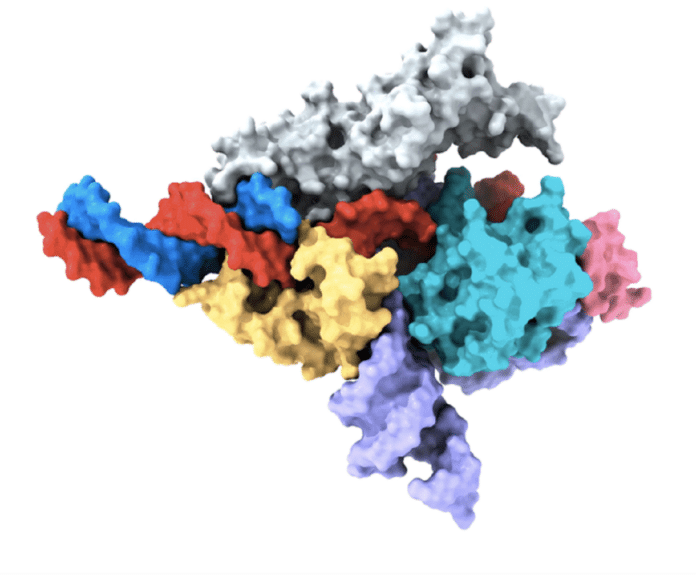
In a remarkable scientific breakthrough, a team led by Feng Zhang from the McGovern Institute for Brain Research at MIT and the Broad Institute of MIT and Harvard has unveiled the world’s first programmable RNA-guided system in eukaryotes. Published in Nature, their study introduces the utilization of a protein named Fanzor as the foundation of this groundbreaking system. By harnessing RNA as a guiding mechanism, Fanzor proteins exhibit extraordinary accuracy in targeting DNA, enabling precise genome editing in human cells.
The compact nature of Fanzor systems also promises easier delivery to cells and tissues as therapeutics, surpassing the challenges faced by CRISPR-Cas systems. Further enhancements to their targeting efficiency could establish Fanzor-based technology as a transformative tool for human genome editing. This study not only expands our understanding of RNA-guided DNA-cutting mechanisms but also reveals their ubiquitous presence across all domains of life, transcending the boundaries of prokaryotes and extending into eukaryotes, including fungi, plants, and animals.
Zhang, senior author of the study, the James and Patricia Poitras Professor of Neuroscience in the MIT departments of Biological Engineering and Brain and Cognitive Sciences, an investigator at MIT’s McGovern Institute, a core institute member at the Broad Institute, and a Howard Hughes Medical Institute investigator say, “CRISPR-based systems are widely used and powerful because they can be easily reprogrammed to target different sites in the genome. This new system is another way to make precise changes in human cells, complementing the genome editing tools we already have.”
The Zhang lab, dedicated to advancing genetic medicines, embarked on a quest to explore RNA-programmable systems beyond CRISPR, seeking new possibilities for targeted gene modulation. Following the discovery of prokaryotic OMEGAs, RNA-programmable systems linked to “jumping genes,” the lab drew parallels between OMEGA systems and Fanzor proteins found in eukaryotes. In their latest study, the researchers expanded their investigation by isolating Fanzors from various organisms, including fungi, algae, amoebas, and the northern quahog clam.
Co-first author Makoto Saito led the characterization of Fanzor proteins, uncovering their role as DNA-cutting endonuclease enzymes guided by ωRNAs, a type of non-coding RNA. This study marks the first identification of this mechanism in animals, unveiling a novel aspect of eukaryotic RNA-guided systems.
In contrast to CRISPR proteins, Fanzor enzymes are encoded within transposable elements in the eukaryotic genome. Phylogenetic analysis conducted by the team indicates that Fanzor genes have undergone horizontal gene transfer from bacteria to eukaryotes. The researchers propose that these OMEGA systems, which predate CRISPR, can migrate between prokaryotes and eukaryotes due to their abundance and ancestral nature. Makoto Saito highlights the significance of these findings, emphasizing the frequent exchange of these proteins across different domains of life.
n their quest for an efficient genome editing tool, the researchers successfully demonstrated Fanzor’s potential by generating targeted insertions and deletions in human cells without causing collateral damage. Although initially less efficient than CRISPR-Cas systems, the Fanzor system’s activity was enhanced through systematic protein engineering, resulting in a tenfold increase in efficiency. Unlike some CRISPR systems, the team found that a fungal-derived Fanzor protein did not exhibit collateral activity, indicating its precision in DNA cleavage without degrading nearby DNA or RNA.
Structural analysis revealed similarities between Fanzor and CRISPR-Cas12 proteins, with the ωRNA playing a significant role in catalytic reactions. With its re-programmability and potential for research and therapeutic applications, Fanzor holds promise as a powerful genome editing technology. The study also highlights the vast diversity of RNA-programmable systems and the researchers’ ongoing exploration to uncover more of these systems in nature.
Journal reference:
- Saito, M., Xu, P., Faure, G. et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature (2023).DOI: 10.1038/s41586-023-06356-2