Protein sequencing refers to methods for determining the amino acid sequence of proteins (or peptides) and analysis of the sequence. Efforts to sequence single protein molecules in nanopores have been hampered by the absence of methods with adequate affectability to observe the molecular differences among all twenty amino acids.
Scientists now demonstrated that nanopores could be used to identify all 20 amino acids in proteins, a significant step toward protein sequencing.
DNA is made up of a double helix of two complementary strands. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, a process referred to as semiconservative replication. For proteins, there is no such natural machinery by which to make copies or to read them.
Adding to the difficulty, 20 amino acids make up proteins, as compared with the four bases in DNA, and numerous small modifications can be made to each amino acid during protein production and folding.
Aleksei Aksimentiev, a co-leader of the study, said, “Many amino acids are very similar. For example, if you look at leucine and isoleucine, they have the same atoms, the same molecular weight, and the only difference is that one structure is the mirror image of the other.”
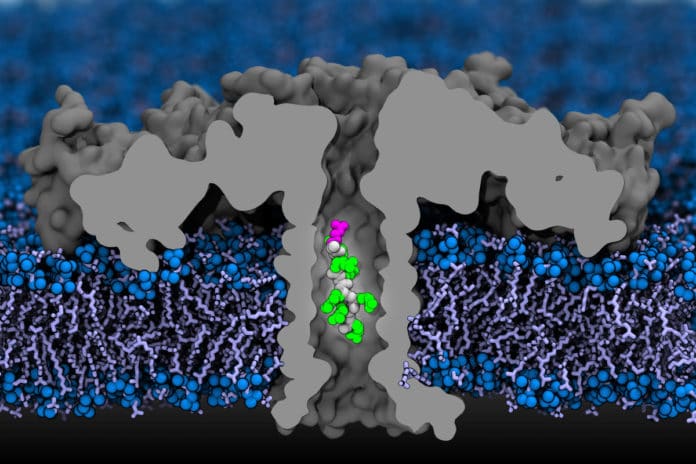
Nanopores, small protein channels embedded in a membrane, are a popular tool for DNA sequencing. Previously, scientists thought that the differences in amino acids were too small to register with nanopore technology. The new study shows otherwise.
Scientists used a membrane channel composed of aerolysin bacteria as their nanopore. In both computer modeling and experimental work, they chopped up proteins and utilized a chemical carrier to drive the amino acids into the nanopore. The carrier molecule additionally kept the amino acids inside the pore long enough for it to enlist a quantifiable distinction in the electrical signature of each amino acid– even leucine and isoleucine, the identical twins.
Abdelghani Oukhaled, a professor of biophysics at Cergy-Pontoise whose team carried out much of the experimental work, said, “This work builds confidence and reassures the nanopore community that protein sequencing is indeed possible.”
Scientists now could differentiate modified types of amino acids by utilizing a more sensitive measurement apparatus or by treating the protein with a chemical to improve differentiation. The estimations are sufficiently exact to distinguish many changes possibly, and significantly more might be perceived by tweaking the pore.
Aksimentiev said, “This is a proof-of-concept study showing that we can identify the different amino acids. The current method for protein characterization is mass spectrometry, but that does not determine the sequence; it compares a sample to what’s already in the database. Its ability to characterize new variations or mutations is limited. With nanopores, we finally could look at those modifications which have not yet been studied.”
“The aerolysin nanopore could be integrated into standard nanopore setups, making it accessible to other scientists. The researchers are now exploring approaches to read the amino acids in sequential order as they are cut from the protein. They also are considering other applications for the system.”
“One potential application would be to combine this with immunoassays to fish out proteins of interest and then sequence them. Sequencing them will tell us whether they’re modified or not, and that could lead to a clinical diagnostic tool.”
“This work shows that there’s no limit to how precisely we can characterize biological molecules. Very likely, one day, we will be able to tell the molecular makeup of the cell – what we are made of, down to the level of individual atoms.”