Chromatin is a dynamic structure that undergoes significant structural changes during DNA replication, transcription, and repair.
Regulation of histone proteins allows the DNA strands to become more tightly or loosely coiled during the process of DNA replication and gene expression. In any case, issues may emerge when histones bunch together or when DNA strands interweave. No doubt, the misregulation of chromatin structures could lead to aberrant gene expression and can eventually prompt developmental disorders or cancers.
Histone chaperones are those proteins that play a vital role in the assembly and disassembly of chromatin. ATAD2 (also termed ANCCA) is a histone chaperone that has been implicated in nucleosome density regulation by histone H3–H4 loading or removal. It is highly overexpressed in various cancers and associated with poor patient prognoses.
Thus, there has been a demand for the development of therapeutic agents, targeting ATAD2 protein, and some clinical trials are already underway. Yet, until now, no specific information about the structure and function of ATAD2 gene have been revealed to the public.
Now, scientists at the UNIST have used cryo-electron microscopy (Cryo-EM) and determined the structure and mechanism of ATAD2 proteins. They presented cryo-EM structures of an ATAD2 family ATPase to atomic resolution in three different nucleotide states.
The study uncovers unique structural features required for histone loading on DNA, and directly visualize the transitions of Abo1 from an asymmetric spiral (ATP-state) to asymmetric ring (ADP- and apo-states) using high-speed atomic force microscopy (HS-AFM).
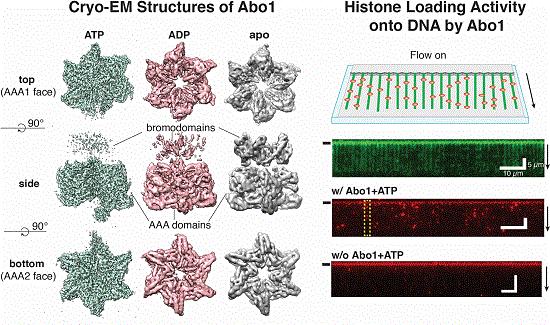
What’s more, the study suggests that the acidic pore of ATP-Abo1 ties a peptide substrate, which is reminiscent of a histone tail. In light of these outcomes, scientists proposed a model whereby Abo1 encourages H3–H4 loading by using ATP.
Professor Ja Yil Lee (School of Life Sciences, UNIST) said, “This study is meaningful, as it reveals the structure and mechanism of histone chaperones protein through the use of cutting-edge techniques in biophysical, such as Cryo-EM. This will accelerate the development of drug candidates, targeting ATAD2.”
The findings of this research were published in Nature Communications on December 17, 2019. This study has been supported through the Mid-career Research Program, funded by the Ministry of Science and ICT (MSIT).
