Using nuclear magnetic resonance (NMR) spectroscopy, MIT chemists have discovered a protein structure that can pump toxic molecules out of bacterial cells. These types of proteins help bacteria become resistant to multiple antibiotics.
Bacteria use many strategies to evade antibiotics. One of them is pumping drugs out through their cell membranes. For several years, scientists have been studying a membrane-bound protein called EmrE. This protein can transfer many different toxic molecules.
The protein EmrE belongs to the group of proteins called the small multidrug resistance (SMR) transporters. The SMR transporters have high sequence conservation across critical regions of the protein.
Several years ago, a technique was demonstrated using NMR to measure the distances between fluorine probes and hydrogen atoms in proteins. Through the technique, scientists were able to determine the structure of a protein as it binds to a molecule that contains fluorine.
Scientists spent several years studying how EmrE transports a drug-like molecule, or ligand called F4-TPP+, across the phospholipid membrane.
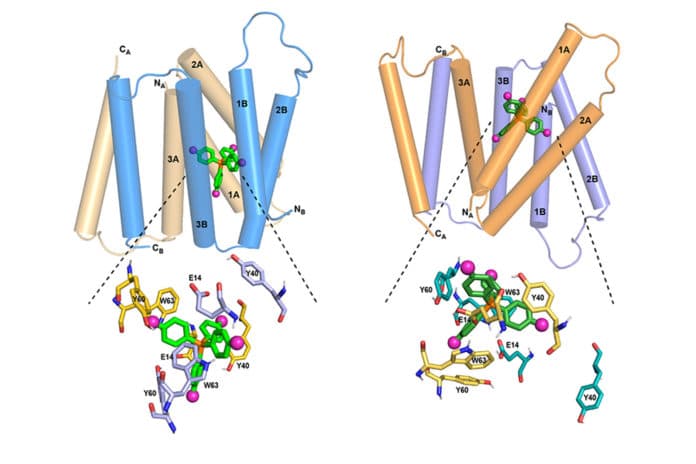
In this new study, MIT chemists used this ligand with the NMR technique to determine an atomic-resolution structure of EmrE. Emre molecule was already known to have four transmembrane helices roughly parallel.
Two EmrE molecules assemble into a dimer so that eight transmembrane helices form inner walls that interact with the ligand as it moves through the channel. Previous studies have revealed the overall topology of the helices, but not of the protein side chains that extend into the channel interior, which are like arms that grab the ligand and help guide it through the channel.
EmrE transports toxic molecules from inside a bacterial cell at neutral pH to the acidic outside. Last year, scientists determined the protein structure as it binds to F4-TPP+ in an acidic environment. In this new study, scientists analyzed the structure at a neutral pH, allowing them to determine how the protein structure changes as the pH changes.
The channel-forming four helices tend to parallel one another at neutral pH. This creates a gateway for the ligand to enter. Dropping pH makes the helices tilt so that the channel is more open toward the outside of the cell. This helps to push the ligand out of the channel.
At the same time, several rings found in the protein side chains shift their orientation in a way that also helps to guide the ligand out of the channel.
The acidic end of the channel is also more welcoming to protons, which enter the channel and help it open further, allowing the ligand to exit more easily.
Mei Hong, an MIT professor of chemistry, said, “Knowing the structure of the drug-binding pocket of this protein, one might try to design competitors to these substrates so that you could block the binding site and prevent the protein from removing antibiotics from the cell.”
“This paper completes the story. One structure is not enough. You need two to figure out how a transporter can open to both sides of the membrane because it’s supposed to pump the ligand or the antibiotic compound from inside the bacteria out of the bacteria.”
Journal Reference:
- Shcherbakov, A.A., Speaker, P.J., Dregni, A.J. et al. High-pH structure of EmrE reveals the mechanism of proton-coupled substrate transport. Nat Commun 13, 991 (2022). DOI: 10.1038/s41467-022-28556-6