A long-running national collaboration has created a detailed atlas of the genome that reveals the location of hundreds of thousands of potential regulatory regions. The atlas interprets as the human genome’s instruction manual- a resource that will help all human biology research moving forward.
Of the three billion base pairs in the human genome, just 2% code for the proteins that manufacture and maintain our bodies. The other 98% harbors, in addition to other things, possible regulatory regions– sequences that give cells the instructions and tools expected to transform protein recipes into an astonishingly complex organism. However, regardless of their significance and predominance, non-coding regions have been studied considerably less than gene-coding sequences, in part because it is more challenging to do so.
The National Human Genome Research Institute launched the Encyclopedia of DNA Elements (ENCODE) collaboration to develop the tools and expertise needed to shed light on our genome’s mysterious majority. Now in its final year, ENCODE has made tremendous advances thanks to the combined scientific and technological prowess of several hundred researchers at dozens of institutions.
Len Pennacchio, a senior scientist at Lawrence Berkeley National Laboratory (Berkeley Lab), said, “We’ve sequenced the human genome, and we largely know where genes are. But when you get outside genes, mapping the function of genomic ‘dark matter’ is much more daunting. It’s a big step forward for us to know how to find the areas within the 98% that are functionally important.”
Along with the original research, scientists also offered technical expertise and materials to other ENCODE consortium teams.
Pennacchio said, “the project’s recent advances will be particularly useful for scientists studying diseases. When trying to determine the underlying causes of a condition, researchers search for genetic variants carried by affected individuals. Sometimes, they find associations with sequences within genes. Still, often the analyses will pinpoint an area that’s far away from any protein-coding sequence, and it isn’t readily apparent what that DNA does. Is it important in the heart or the stomach? Is it important all the time or just at certain phases of development?”
“Our datasets give scientists clues as to when and where that sequence functions, and which gene or genes it affects. It gives you an immediate path to follow to learn more, where previously we’d have few hints.”
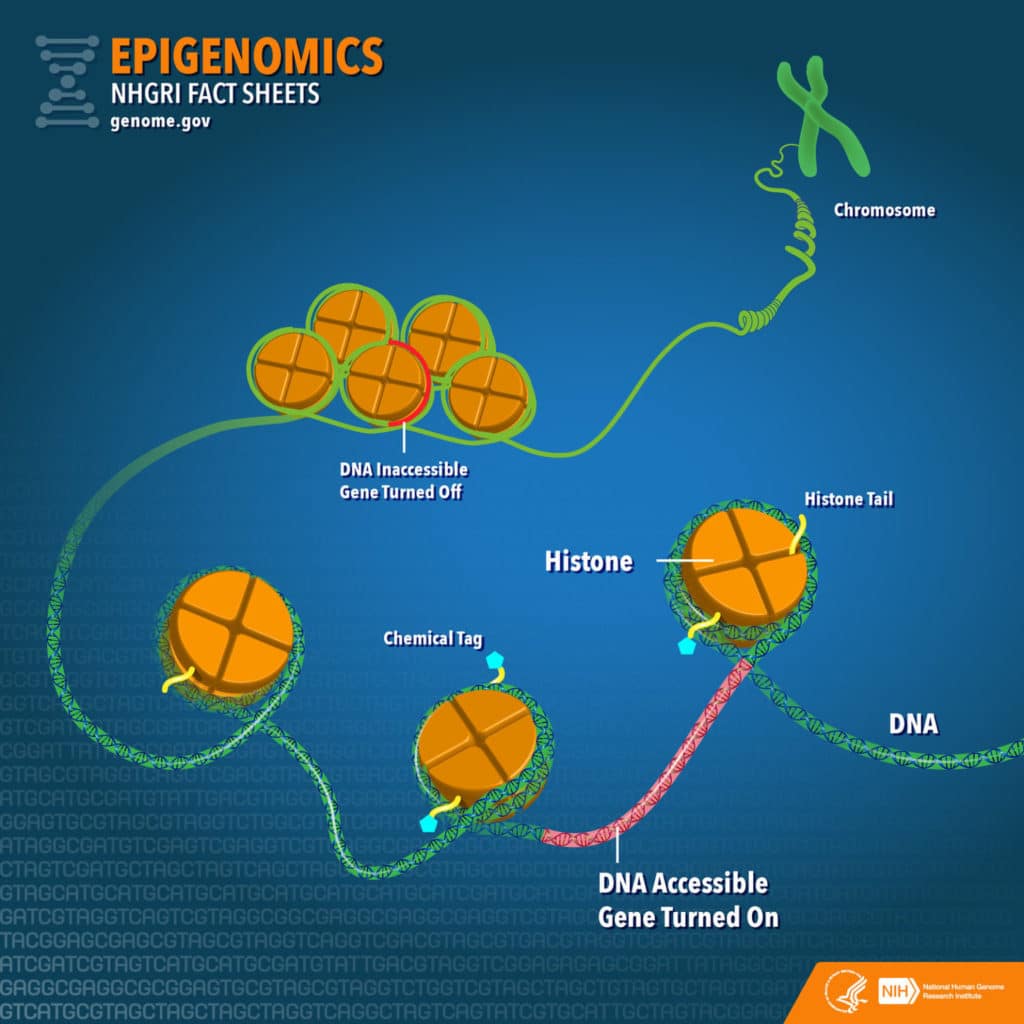
In past phases, scientists identified all DNA sequences that regulate gene expression. They also established how different regions of our chromosomes are modified and stored. Their information reveals a great deal about how cells can express or silence genes differently depending on timing and where they are located in the body. The earlier work was mostly performed on DNA extracted from human cell lines.
Axel Visel, also a Berkeley Lab senior scientist, said, “Thanks to ENCODE 2, we had a pretty good map of how DNA is modified along the genome, but what was missing was the legend for that map.”
“ENCODE phase 3 has been all about understanding what these different modifying marks we found in cell lines mean in terms of a real organism.”
For the phase 3 experiments, the Berkeley Lab group, along with numerous other ENCODE consortium teams, began applying their analyses to mouse tissues, as the mouse genome is very similar to ours and many of the DNA modifications and on-off switches for gene expression are known to be the same.
The Berkeley Lab team, which has been involved in the project for 12 years, played an especially significant role in ENCODE 3. They are renowned leaders in the use of ChIP-seq, a technique that allows scientists to locate transcription factors and modified proteins on chromatin (the densely packed state that DNA exists in when not activated for transcription or replication), and then to analyze how these molecules are interacting with the sequences. They are also known for their expertise in transgenic assays, a technique used to test if potential gene switches function as predicted.
Working closely with Bing Ren at the Ludwig Institute for Cancer Research, the team used ChIP-seq to study the changing landscape of chromatin in embryonic mice and then carried out hundreds of transgenic assays to validate these findings. After thousands of experiments, they generated a dataset covering diverse body tissues at eight developmental stages, significantly expanding the scientific community’s knowledge of DNA dynamics during mouse development and creating a resource for biomedical researchers seeking to learn more about human evolution.
Dickel said, “Over the years, we’ve worked extensively with the other groups that were involved in ENCODE and built great complementary relationships. This is the kind of progress that comes from good collaborations, rather than competition.”
4 of the 15 new ENCODE papers published this week as part of a special collection in Nature.