Using single-cell RNA sequencing, scientists at the Yale School of Medicine (YSM) are learning single-cell RNA sequencing of infected human bronchial epithelial cells (HBECs) to determine how the virus infects and alters healthy cells.
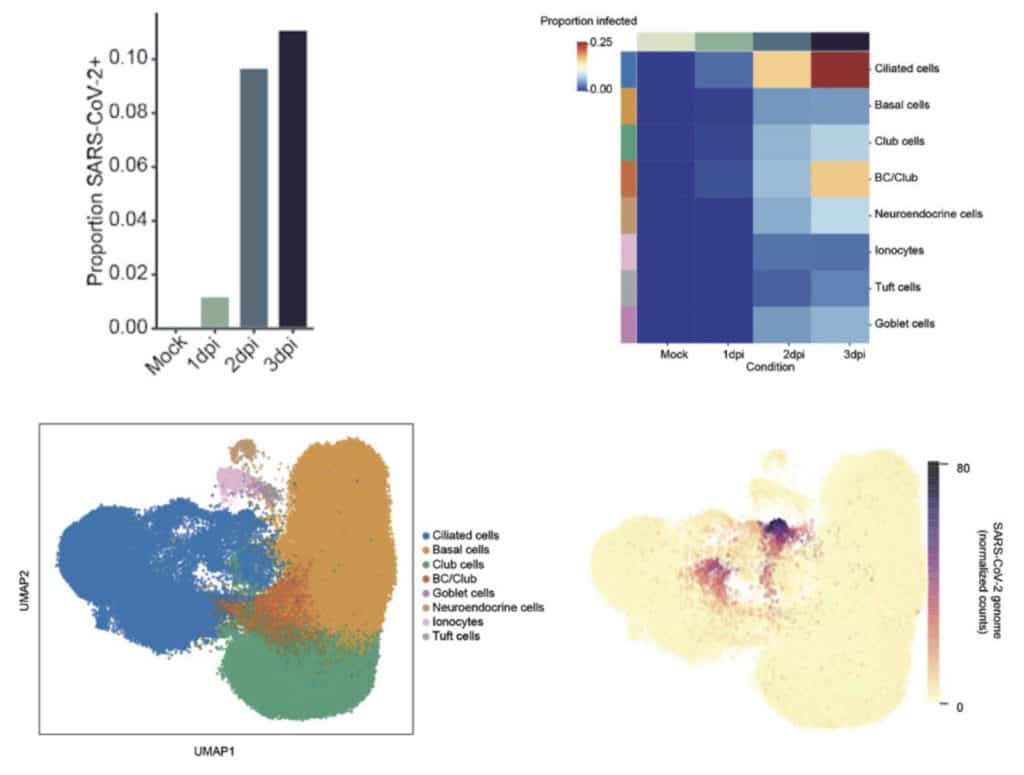
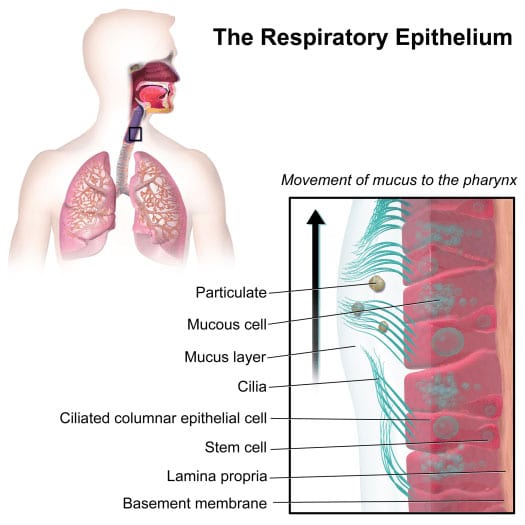
Scientists mainly identified ciliated cells as the major target of SARS-CoV-2 infection. The bronchial epithelium acts as a defensive barrier against allergens and pathogens. Cilia expels mucus and different particles from the respiratory tract. Their discoveries offer knowledge into how the virus turns deadly.
For their study, scientists tainted HBECs in an air-liquid interface with SARS-CoV-2. Over three days, they utilized single-cell RNA sequencing to distinguish signatures of infection dynamics, for example, the number of tainted cells across cell types, and whether SARS-CoV-2 enacted an immune response in contaminated cells. Van Dijk, who has practical experience in single-cell advances, used propelled calculations to create working speculations.
David van Dijk, Ph.D., an assistant professor of medicine in the Section of Cardiovascular Medicine and Computer Science, utilized advanced algorithms to develop working hypotheses.
Post-doctoral associate Mia Alfajaro, Ph.D., said, “Machine learning allows us to generate hypotheses. It’s a different way of doing science. We go in with a few hypotheses as possible. Measure everything we can measure, and the algorithms present the hypothesis to us.”
Using electron microscopy, scientists then observed the structural basis of the virus and target cells. These observations provide insights about host-virus interaction to measure SARS-CoV-2 cell tropism, or the ability of the virus to infect different cell types, as identified by the algorithms. Three days later, several cultured cells became tainted.
Scientists analyzed data from the infected cells along with neighboring bystander cells. They observed ciliated cells were 83% of the infected cells. These cells were the first and primary source of infection throughout the study. The virus also targeted other epithelial cell types, including basal and club cells. The goblet, neuroendocrine, tuft cells, and ionocytes were less likely to become infected.
The gene signatures uncovered an innate immune response-related to a protein called Interleukin 6 (IL-6). The examination additionally indicated a shift in the polyadenylated viral transcripts. In conclusion, the (uninfected) bystander cells likewise demonstrated an insusceptible response, likely because of signs from the contaminated cells. Pulling from a vast number of genes, the algorithms locate the genetic differences between infected and non-infected cells.
Scientists are planning to continue this study to further examine the severity of SARS-CoV-2 compared to other types of coronaviruses, and conduct tests in animal models.
Other co-authors of the study include Craig Wilen, MD, Ph.D., Neal Ravindra, Ph.D., Mia Madel Alfajaro, Ph.D., and Victor Gasque. This study was also made possible by several collaborators at YSM, including Jin Wei, Renata Filler, Nicholas Huston, Han Wan, Klara Szigeti-Buck, Bao Wang, Ruth Montgomery, Stephanie Eisenbarth, Anna Marie Pyle, Akiko Iwasaki, Tamas Horvath, and Ellen Foxman.
Journal Reference:
- Neal G. Ravindra, Single-cell longitudinal analysis of SARS-CoV-2 infection in human bronchial epithelial cells. DOI: 10.1101/2020.05.06.081695