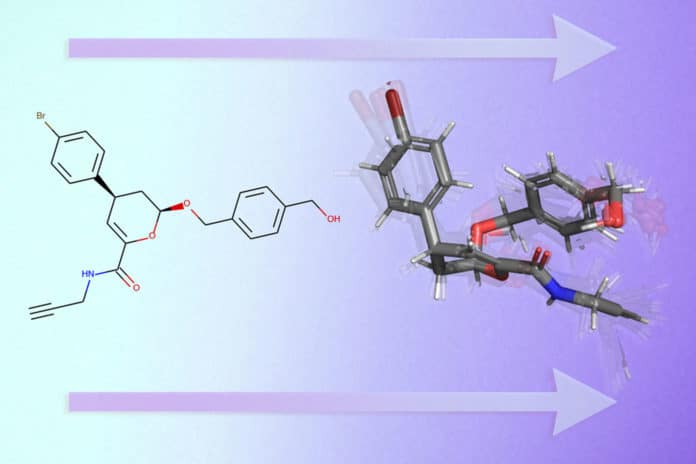
Dealing with molecules in their natural 3D structure is essential in cheminformatics or computational drug discovery. These 3D conformations determine the biological, chemical, and physical properties.
Determining the 3D shapes of a molecule helps understand how it will attach to specific protein surfaces. But, that’s not an easy task. Plus, it is time consuming and expensive process.
MIT scientists have come up with a solution to ease this task. Using machine learning, they have created a deep learning model called GeoMol that predicts the 3D shape. As molecules are generally represented in small graphs, the GeoMol works based on a graph in 2D of its molecular structure.
Unlike other machine learning models, the GeoMol processes molecules in only seconds and performs better. Plus, it determines the 3D structure of each bond individually.
Usually, pharmaceutical companies need to test several molecules in lab experiments. According to scientists, the GeoMol could help those companies accelerate the drug discovery process by diminishing the need for testing molecules.
Lagnajit Pattanaik, a graduate student in the Department of Chemical Engineering and co-lead author of the paper, said, “When you are thinking about how these structures move in 3D space, there are really only certain parts of the molecule that are flexible, these rotatable bonds. One of the key innovations of our work is that we think about modeling conformational flexibility like a chemical engineer would. It is really about trying to predict the potential distribution of rotatable bonds in the structure.”
GeoMol leverages a recent tool in deep learning called a message passing neural network. It is specially designed to operate on graphs. By adapting a message passing neural network, scientists could predict specific elements of molecular geometry.
The model, at first, predicts the lengths of the chemical bonds between atoms and the angles of those individual bonds. The arrangement and connection of atoms determine which bonds can rotate.
It then predicts the structure of each atom’s surrounding individually. Later, it assembles neighboring rotatable bonds by computing the torsion angles and then aligning them.
Pattanaik said, “Here, the rotatable bonds can take a huge range of possible values. So, using these message passing neural networks allows us to capture a lot of the local and global environments that influence that prediction. The rotatable bond can take multiple values, and we want our prediction to be able to reflect that underlying distribution.”
As mentioned above, the model determines each bond’s structure individually; it explicitly defines chirality during the prediction process. Hence, there is no need for optimization after-the-fact.
Octavian-Eugen Ganea, a postdoc in the Computer Science and Artificial Intelligence Laboratory (CSAIL), said, “What we can do now is take our model and connect it end-to-end with a model that predicts this attachment to specific protein surfaces. Our model is not a separate pipeline. It is very easy to integrate with other deep learning models.”
Scientists used a dataset of molecules and the likely 3D shapes they could take to test their model. By comparing the model with other methods and models, they evaluated how many were likely to capture 3D structures. They found that GeoMol outperformed the other models on all tested metrics.
Pattanaik said, “We found that our model is super-fast, which was exciting to see. And importantly, as you add more rotatable bonds, you expect these algorithms to slow down significantly. But we didn’t see that. The speed scales nicely with the number of rotatable bonds, which is promising for using these types of models down the line, especially for applications where you are trying to predict the 3D structures inside these proteins quickly.”
Scientists are planning to use GeoMol in high-throughput virtual screening. This would help them determine small molecule structures that interact with a specific protein.
Journal Reference:
- Octavian-Eugen Ganea, Lagnajit Pattanaik, Connor W. Coley, Regina Barzilay, Klavs F. Jensen, William H. Green, Tommi S. Jaakkola. GeoMol: Torsional Geometric Generation of Molecular 3D Conformer Ensembles. arXiv:2106.07802