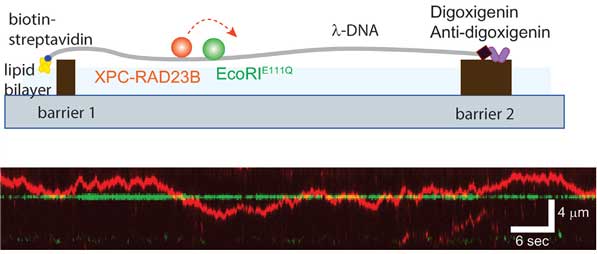
Scientists at the UNIST have recently identified the movement of human NER protein, XPC-RAD23B, that plays a vital role in detecting DNA damage in our bodies. Using a single-molecule spectroscopy technique called DNA curtain, scientists found that the protein moved along the DNA and identified the damage.
Na Young Cheon, the first author of the study, said, “The XPC-RAD23B was able to pass through other proteins easily, so it was able to find damage to up to 3 billion DNA sites quickly. This will enable us to develop molecular biological methods to identify DNA damage that causes various diseases quickly.”
DNA containing genetic information can be easily damaged and altered by UV or toxic substances. These changes don’t have mutations because of the nucleotide ablation repair (NER), which rapidly and precisely reestablishes damaged DNA in our bodies, proceeds.
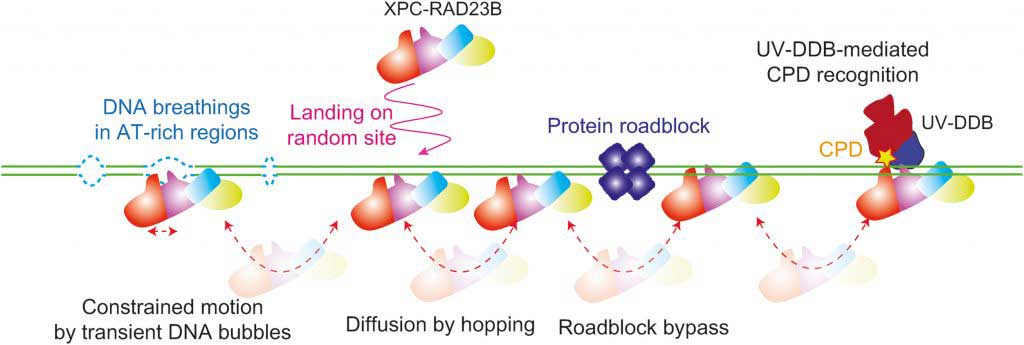
This fix is the interaction of different proteins; the starting point for the XPC-RAD23B protein is to identify the site of damage.
Scientists believe that the study will serve as a molecular biological basis for treating a variety of genetic diseases, including skin cancer and pigmentary dry skin caused by DNA damage.
Professor Lee said, “Current microscopic techniques do not allow us to accurately observe the interaction of proteins and DNA in the cell nucleus. It provides the foundation for observing more precisely the interaction of DNA and protein that causes various diseases.”
The Mid-career Research Program has supported this study through the National Research Foundation (NRF), the POSCO Science Fellowship of POSCO TJ Park Foundation, and the Korean Institute for Basic Science (IBS). It has also been featured as NAR’s Breakthrough Articles.
The findings are published in the journal, Nucleic Acids Research (IF 11.56) on August 2, 2019.
