Protein design provides an opportunity to test our understanding of protein folding and investigate how amino acid sequences determine unique folded structures. In previous studies, scientists decided to design small versions of ‘ideal’ proteins, which are structures without internal energetic frustration.
These proteins are designed with beta-strands, a molecular feature that acts as a critical structural role for the molecules.
Despite being structurally ideal, these proteins are, however, too small to harbor functional sites.
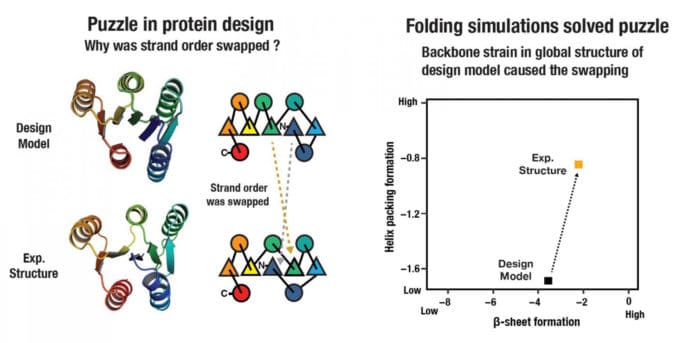
Hence, in a new study, scientists set out to test the generality of the design principles by applying them to the design of larger alpha-beta proteins with five and six beta-strands. And the outcomes were surprising: their experimental structures differed from their computer models, resulting in proteins that folded differently by swapping the internal locations of their beta-strands.
A team from Japan and the United States struggled with the strand swapping puzzle, but they concluded by iterating between computational design and laboratory experiments.
That strand swapping takes place due to the strain of the whole system on the foundational backbone structure.
Co-first author Gaohua Liu, chief scientific officer of Nexomics Biosciences, said, “We emphasize that experimental structure determination is important for iterative improvement of computational protein design.”
Gaetano Montelione, co-author and professor of chemistry and chemical biology at Rensselaer Polytechnic Institute, said, “Sometimes we learn the most from these ideal proteins when their experimental structures differ, rather than match, their intended design, since this can lead to a deeper understanding of the underlying principles.”
According to Nobuyasu Koga, the strain is global instead of connection to connection. Proteins can adjust the length and register of strands across the system to alleviate this backbone strain.
David Baker, co-author and professor of biochemistry at the University of Washington, said, “We would like to design proteins with more complex functional sites by incorporating non-ideal features such as longer loops, which are important not only for function but also for relieving global backbone strain.”
Journal Reference:
- Koga, N., Koga, R., Liu, G. et al. Role of backbone strain in de novo design of complex α/β protein structures. Nat Commun 12, 3921 (2021). DOI: 10.1038/s41467-021-24050-7