Coronaviruses (CoVs), enveloped positive-sense RNA viruses, are characterized by club-like spikes that project from their surface, an unusually large RNA genome, and a unique replication strategy.
Precisely how coronavirus replicate is a complex puzzle with many missing pieces. Also, in the era of this pandemic, understanding it has become a matter of acute urgency.
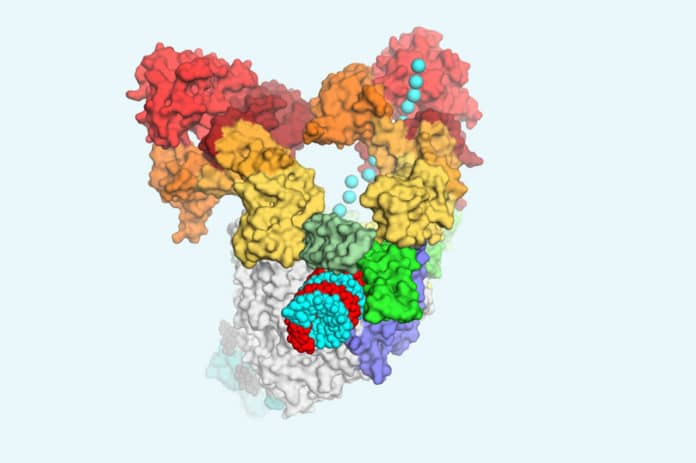
In a new study, Rockefeller scientists provide a crucial piece of the puzzle: an atomic-level resolution view of SARS-CoV-2‘s replication system. Scientists now have an additional structural template that can help drug developers discover new compounds that could get into this molecular machine and make it stop.
The coronavirus copies its genetic material with the help of an enzyme: the RNA-dependent RNA polymerase, or RdRp. As it is essential to viral replication, this machinery is thought to be a promising target for antiviral drugs. Some existing antivirals and several new candidates under investigation specifically for COVID-19 act on RdRp—including remdesivir, which is currently being used in several countries for the treatment of severe cases.
These antiviral medications attempt to hold up into nooks and crannies of the giant RdRp molecule, similar to clogs inside its gears, carrying the machine to an end. To pull this off, a compound should be exceptionally exact.
This is why scientists are trying to design a successful compound that needs the most detailed picture of the RdRp they can get.
James Chen, a postdoctoral associate in Seth Darst’s lab and one of the study’s first authors, said, “Further complicating matters is that RdRp doesn’t work alone. It joins hands with several other proteins, including another crucial enzyme called the helicase, which in its own right, is a promising target for COVID-19 drug discovery. This tight cluster of RdRp and associated proteins is likely what the enzyme looks like outside the lab and in its native environment, inside an infected cell.”
In this study, scientists used a powerful imaging technique called cryo-electron microscopy to see what precisely this multipart machine looks like.
They found that even when they form a complex, the cavities of the RdRp or the helicase don’t change shape, so molecules designed to inhibit these enzymes in isolation may still work on the duo.
What’s more, the image reveals several previously unknown sites in the machine that may be vulnerable to drugs—including one spot at the interface between the two enzymes, a joint that could be potentially dismantled by an interfering molecule.
The unprecedented resolution at which the team produced their 3-D map of the RdRp-helicase complex will aid computational studies in which researchers explore the function of drug candidates “virtually,” based on their knowledge of the chemical structure of the molecules.
Brandon Malone, a graduate student at Rockefeller and the study’s co-author, said, “When one is looking for molecules that can lodge into a particular binding pocket, having a detailed picture of what it looks like greatly improves the computational docking precision.”
Chen said, “Now we’ll be able to not only propose models for the mechanics of viral replication but also test those models.”
Journal Reference:
- James Chen et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex, Cell (2020). DOI: 10.1016/j.cell.2020.07.033