Purdue University, Super-resolution microscopy is a technique that can “see” beyond the diffraction limit of light. The system has accumulated expanding interest as of late, particularly since its designers won the Nobel Prize in Chemistry in 2014.
By exploiting fluorescence, super-resolution microscopy presently enables researchers to observe cells and their inside structures and organelles in a way at no other time conceivable.
Huge numbers of the molecular complexes inside cells are comprised of various proteins. Since current procedures of super-resolution microscopy ordinarily can just utilize a couple of fluorescent colors, it is hard to observe distinctive proteins and unravel the complex architecture and hidden assembly of the cells inside structures.
A much more prominent test is to beat the noise inherent to the super-resolution techniques and fluorescent labeling, to accomplish the full resolution potential.
Researchers from the lab of Suliana Manley at EPFL have now tackled both issues by building up another technique to break down and recreate super-resolution pictures and re-align them in a way that various proteins can be put inside a single 3D volume.
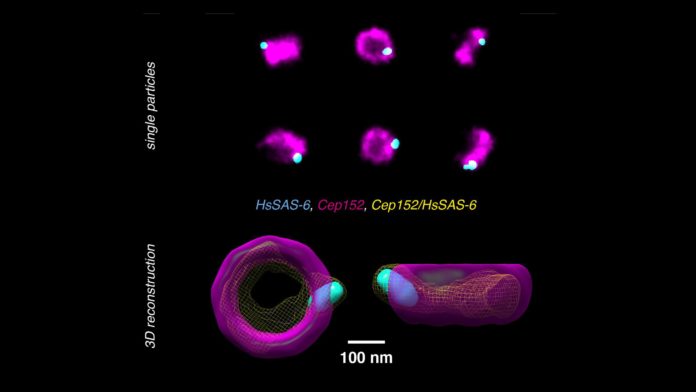
The technique works with pictures taken with large field-of-view super-resolution microscopy, with each picture containing hundreds of two-dimensional projections of a labeled structure in parallel.
Each 2D see represents a marginally unique orientation of the structure so that with a dataset of thousands of views, the strategy can computationally remake and align the 2D pictures into a 3D volume.
By combining data from an expansive number of single images, the noise is diminished and the compelling goals of the 3D reconstruction are improved.
With the assistance of Pierre Gönczy’s lab at EPFL, the specialists tried the strategy on human centriole complexes. Centrioles are sets of cylindrical and hollow atomic congregations that are pivotal in helping the cell divide. Utilizing the new multicolor super-resolution recreation strategy, the specialists could reveal the 3D engineering of four proteins critical for centriolar assembly amid organelle biogenesis.
Suliana Manley said, “With this method if the proteins in the structure can be labeled, there is no limit to the number of colors in the 3D reconstruction. Plus, the reconstruction is independent of the super-resolution method used, so we expect this analysis method and software to be of broad interest.”
The study is published in the journal Nature Methods.