Our body organs constantly communicate with each other. These communication networks play a critical role in our bodies every day. However, it isn’t easy to uncover such pathways.
To label and track the protein signals that enable organ-to-organ communication, scientists at Scripps Research, the University of Southern California, and elsewhere have successfully created a model that could shape our molecular understanding of healthy versus diseased tissue. It also shapes our understanding of inter-organ communication’s role in disease onset and progression.
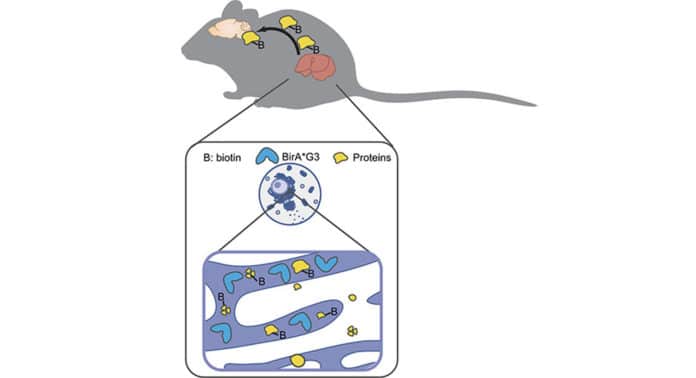
The new mouse model marks the proteins a cell secretes and tracks their movement throughout the body.
Study co-first author Ilia Droujinine, Ph.D., Scripps fellow and principal investigator in the Department of Molecular Medicine at Scripps Research said, “This new model can be likened to establishing a passport system in the body, as we are identifying where proteins are coming from and where they’re going. We can finally bring these interconnected communications networks to light, and then develop treatments based on this new knowledge.”
Using other methods such as viral approaches, scientists understood protein secretion and how organs communicate with each other. These methods offer significant insights into the proteins expressed in an organism. Though, they are not sensitive enough to label low-abundance proteins or the origin and ultimate destination of protein interactions.
But with this new model, scientists can now understand a particular protein’s exact path.
Scientists used an enzyme called BirA*G3. This enzyme labels secreted proteins with a biotin tag, which are then detected in live mice using quantitative mass spectrometry proteomics. This revealed where the proteins originated and where they traveled in the body.
Researchers discovered that all released proteins, including modest levels of proteins with hormone-like characteristics, were successfully tagged when BirA*G3 was widely active throughout the body. The remarkable specificity of the model was also demonstrated when BirA*G3 was selectively activated in the liver, highlighting only secreted proteins associated with that organ system.
Andrew McMahon, Ph.D., senior author of the study and chair of the Department of Stem Cell Biology and Regenerative Medicine at the University of Southern California, said, “Given the central role of key secreted proteins such as insulin, there is a great deal of interest in identifying novel secreted proteins. Genome studies suggest that many new proteins remain to be characterized. We’re looking forward to a deep dive into this area now that we have validated the technology.”
Droujinine said, “There are countless research applications for this technology. With this model, scientists can start to map unexplored disease pathways and ultimately develop targeted treatments, as many diseases originate in a single organ and then eventually spread to others. Cancer, with its metastatic properties, is one example. For another, studies have shown that many health complications arising from obesity could be due to faulty organ communication, yet many molecular mechanisms remain unknown.”
“Any protein we discover that plays a role in disease has the potential to be translated into a therapeutic.”
Journal Reference:
- Rui Yang, Amanda S. Meyer et al. A genetic model for in vivo proximity labeling of the mammalian secretome. Open Biology. August 2022. DOI: 10.1098/rsob.220149