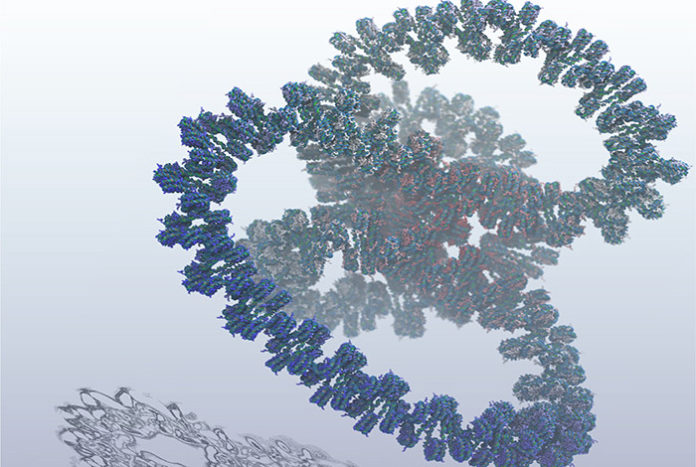
The growing interest in the complexity of biological interactions is continuously driving the need to increase system size in biophysical simulations, requiring not only powerful and advanced hardware but adaptable software that can accommodate a large number of atoms interacting through complex forcefields.
To address this, scientists at the Los Alamos National Laboratory have created the largest simulation to date of an entire gene of DNA, a feat that required one billion atoms to model and will help researchers to better understand and develop cures for diseases like cancer.
Karissa Sanbonmatsu, a structural biologist at Los Alamos said, “It is important to understand DNA at this level of detail because we want to understand precisely how genes turn on and off. Knowing how this happens could unlock the secrets to how many diseases occur.”
Scientists started with modeling genes at the atomistic level to create a complete explanation of how DNA expands and contracts, which controls genetic on/off switching. They then ran the breakthrough simulation on Los Alamos’ Trinity supercomputer, the sixth fastest in the world. The capabilities of Trinity primarily support the National Nuclear Security Administration stockpile stewardship program, which ensures safety, security, and effectiveness of the nation’s nuclear stockpile.
When DNA is more compacted, genes are turned off and when the DNA expands, genes are turned on. Researchers do not yet understand how or why this happens.
While the atomistic model is key to solving the mystery, simulating DNA at this level is no easy task and requires massive computing power.
Anna Lappala, a polymer physicist at Los Alamos said, “Right now, we were able to model an entire gene with the help of the Trinity supercomputer at Los Alamos. In the future, we’ll be able to make use of exascale supercomputers, which will give us a chance to model the full genome.”
Exascale computers are the next generation of supercomputers and will run calculations many times faster than current machines. With that kind of computing power, researchers will be able to model the entire human genome, providing even more insight into how genes turn on and off.
In the new study published in the Journal of Computational Chemistry April 17, the Los Alamos team partnered with researchers from the RIKEN Center for Computational Science in Japan, the New Mexico Consortium and New York University to collect a large number of different kinds of experimental data and put them together to create an all-atom model that is consistent with that data.
Simulations of this kind are informed by experiments, including chromatin conformation capture, cryo-electron microscopy, and X-ray crystallography as well as a number of sophisticated computer modeling algorithms from Jaewoon Jung (RIKEN) and Chang-Shung Tung (Los Alamos).