Some microbes in nature keep us healthy while some make us sick. Scientists from the University of Illinois shows the power of ‘big data’ genome technology by making advancement in bioinformatics. They have developed a tool that searches microbial genomes by identifying clusters of genes.
Scientists have explained how their custom software learns to recognize predictive microbial genomes features.
Scientists said, “With genome sequencing going at the pace it has . . . there’s a dearth of functional information about what these genes are doing. It becomes increasingly important to make sense of and interpret metabolic pathways, especially biosynthetic gene clusters encoded by microbes.”
Scientists particularly were interested in a class of molecules- RiPPs. Because proteins like RiPPs are made from amino acids chains. They are encoded by genes and undergo a chemical change after they are made.
Lead authors Jonathan Tietz said, “RiPPs have some particular advantages compared to other more traditional, classes of natural products. They’re usually larger and more structurally complex. This allows them to interact with cellular machinery in ways a smaller molecule cannot.”
“More points of contact with their cellular targets mean RiPPs can hang on better and perform more complicated tasks. At the same time, despite their complexity, RiPP biosynthesis . . . makes for greater potential for genetic re-engineering of natural products to tailor physical and pharmacological properties.”
Previously, scientists found RiPPs is a potentially useful natural product. But, after long research, scientists revealed a range of products including some RiPPs. For example, the low-hanging fruit has been plucked; searches turn up the same common compounds over and over again. So scientists have found a way to “climb higher” and uncover novel natural products: genome mining.
Scientists actually want to speed up drug discovery by searching through the microbial genomes. By doing this, scientists can greatly increase the odds that they will isolate a compound that has never been seen before. In addition, this method has the ability to show what a group of genes might be capable of producing.
Lead author Christopher Schwalen said, “In a practical sense the question became, is there a better way to harness available genomes for augmenting these discovery pipelines. That’s where we started.”
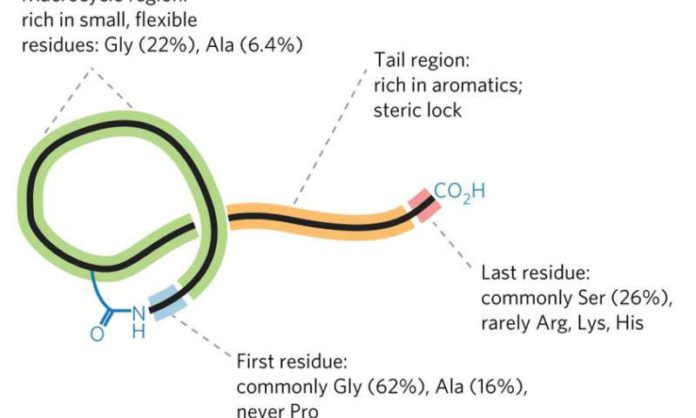
Scientists want the tool to recognize the groups of microbial genomes whose products work together to synthesize a RiPP. They created it by using a class of RiPPs called lasso peptides. The clusters of genes that produce lasso peptides are small and generic-looking. It makes cluster difficult to find even in a manual search.
The tool is named as RODEO. RODEO stands for Rapid Open reading frame Description and Evaluation Online. It detects robustly identified promising gene clusters in a broad array of microbial genomes. In addition, it could customize for searching the gene clusters of other classes of RiPPs as well.
Scientists reported RODEO can find 1300 novel lasso peptides with particularly unusual structures that make them promising as potential therapeutics.
Mitchell said, “If you want to show that you have a useful tool, you pick the hardest example. But, as a chemist, lasso peptides are extremely interesting. Peptides are used as drugs that cannot be given orally because they would be digested.”
“Lasso peptides are different. You can actually boil these, you can throw proteases at them, you can autoclave them and they don’t lose their activity, they are basically a little peptide knot that is extremely resistant to such assaults.”
“We can now use genomic prioritization to find molecules that without any doubt are structurally novel. The challenge is that a useful molecule or not? But the more molecules you can connect to genes, the better informed we’re going to get. So that’s the next 10 years of discovery,” he added.