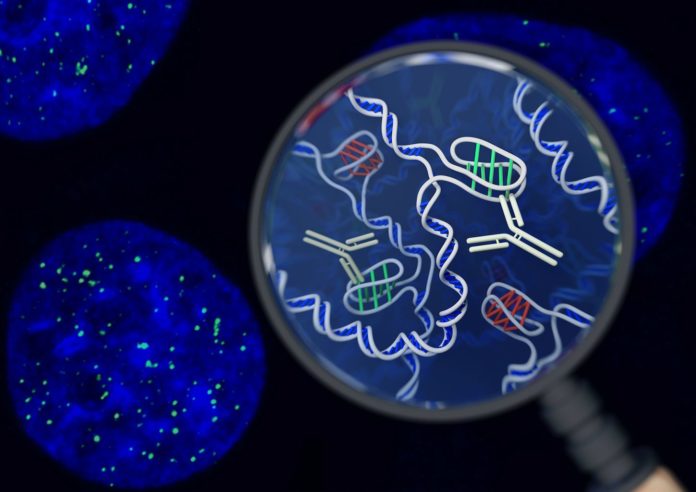
For the first time in the world, UNSW researchers have identified a new DNA structure – called the i-motif, a twisted ‘knot’ of DNA inside cells. Scientists from the Garvan Institute of Medical Research have made this discovery by directly seeing i-motif inside living cells.
Our DNA lies somewhere inside the cells in our body. The data in the DNA code – each of the 6 billion A, C, G and T letters– give exact guidelines to how our bodies are manufactured, and how they work.
The notorious ‘twofold helix’ state of DNA has captured the general population creative energy since 1953 when James Watson and Francis Crick broadly revealed the structure of DNA. In any case, it’s currently realized that short extends of DNA can exist in different shapes, in the lab at any rate – and researchers presume that these diverse shapes may assume an imperative part in how and when the DNA code is ‘read’.
The new shape appears to be altogether unique to the twofold stranded DNA twofold helix.
Associate Professor Daniel Christ, Head of the Antibody Therapeutics Lab at Garvan said, “When most of us think of DNA, we think of the double helix. This new research reminds us that totally different DNA structures exist and could well be important for our cells.”
Associate Professor Marcel Dinger, Head, of the Kinghorn Centre for Clinical Genomics at Garvan, said, “The i-motif is a four-stranded ‘knot’ of DNA. In the knot structure, C letters on the same strand of DNA bind to each other – so this is very different from a double helix, where ‘letters’ on opposite strands recognize each other, and where Cs bind to Gs [guanines].”
To detect the i-motifs inside cells, the researchers developed a precise new tool – a fragment of an antibody molecule – that could specifically recognize and attach to i-motifs with a very high affinity. Until now, the lack of an antibody that is specific for i-motifs has severely hampered the understanding of their role.
With the new instrument, specialists revealed the area of ‘I-motifs’ in a scope of human cell lines. Utilizing fluorescence methods to pinpoint where the I-themes were found, they recognized various spots of green inside the core, which show the situation of I-motifs.
Dr. Mahdi Zeraati, whose research underpins the study’s findings said, “What excited us most is that we could see the green spots – the i-motifs – appearing and disappearing over time, so we know that they are forming, dissolving and forming again.”
The scientists demonstrated that I-motifs for the most part shape at a specific point in the cell’s ‘life cycle’ – the late G1 phase when DNA is as a rule effectively ‘read’. They likewise demonstrated that I-motifs show up in some promoter districts – regions of DNA that control whether qualities are turned on or off – and also in telomeres, the ‘end segments’ of chromosomes that are critical in the aging process.
Dr. Zeraati says: “We think the coming and going of the i-motifs is a clue to what they do. It seems likely that they are there to help switch genes on or off, and to affect whether a gene is actively read or not.”
Associate Professor Christ says: “We also think the transient nature of the i-motifs explains why they have been so very difficult to track down in cells until now.”
Their work is published today in the leading journal Nature Chemistry.