A new DNA modification signature has been recently discovered in the genome of zebrafish. This discovery was made by scientists from the Garvan Institute of Medical Research.
Scientists found that strangely elevated levels of DNA repeats of the sequence ‘TGCT’ in the zebrafish genome go through a modification called methylation. This modification may change the shape or activity of the surrounding DNA.
Dr. Ozren Bogdanovic, who heads the Developmental Epigenomics Lab at Garvan and Senior Research Fellow at the School of Biotechnology and Biomolecular Sciences, UNSW Sydney, said, “We’ve revealed a new form of DNA methylation in zebrafish at TGCT repeats, and crucially, the enzyme that modifies. These findings open the field to new possibilities in studying the epigenome—the additional layer of instructions on DNA that change how genes are read—and understand how it may be clinically relevant.”
The first author of the paper, Ph.D. student Sam Ross said, “DNA methylation is vital to cellular function, as it controls which genes are turned on and off. This is why the cells in our body can carry out vastly different functions, despite having almost identical DNA.”
The DNA of life on Earth naturally stores its information in just four essential chemicals—guanine, cytosine, adenine, and thymine, commonly referred to as G, C, A, and T, respectively. In vertebrates, methylation occurs mostly where the letter G follows a C (‘CG’), but there are some exceptions. One example is methylation at non-CG sites in human brain cells, aberrations linked to Rett Syndrome, a genetic disorder that impairs growth, movement, and speech in children.
To investigate non-CG methylation further, scientists conducted comprehensive profiling of the zebrafish genome. This vertebrate organism is a distant evolutionary relative of humans and shares 70% of our genes, making it a useful model for studying the effects of human genes.
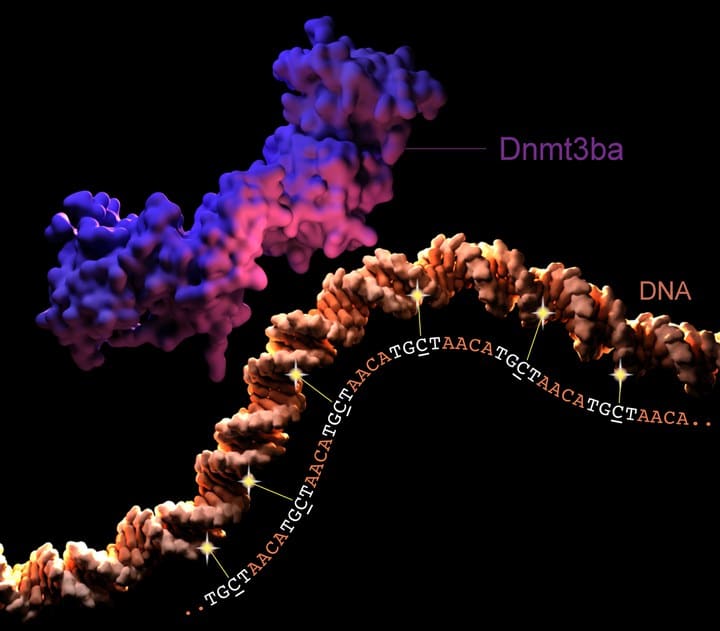
Scientists discovered that methylation occurred where the sequence ‘TGCT’ appeared multiple times, close together.
Dr. Bogdanovic said, “We were fascinated to see that methylation levels at TGCT repeats were higher than any non-CG methylation previously observed in the majority of adult vertebrate tissues. Further, this methylation was present at high sperm and egg levels, absent in the fertilized egg. It then appeared again in the growing embryo, reaching its highest levels in adult tissues such as the brain and gonads. While we are yet to reveal how this modification changes gene expression, we believe TGCT methylation to be linked to the ‘awakening’ of the embryonic genome in zebrafish.”
The enzyme Dnmt3ba was responsible for methylating the TGCT repeats in the zebrafish genome.
Dr. Bogdanovic said, “While it’s unclear if a similar modification occurs in animals more broadly, our discovery in zebrafish is significant because it means we can start to selectively manipulate this atypical form of methylation in a model organism. It means we can change the levels of Dnmt3ba to see what happens when we remove just one form of methylation, but not another.”
“This could greatly facilitate our understanding of how changes in atypical methylation patterns affect specific tissues such as the brain, to gain further insights into the molecular mechanisms of neurodevelopmental disorders.”
“We hope that our findings will help us develop new experimental models that can be used to study epigenetics in a way that has not been possible thus far.”
Journal Reference:
- Samuel E Ross et al., Developmental remodeling of non-CG methylation at satellite DNA repeats, Nucleic Acids Research (2020). DOI: 10.1093/nar/gkaa1135
