In an effort to understand millions of proteins in the brain, scientists from Northwestern University and the University of Pittsburgh have developed a new approach to see how proteins work. As a result, this approach could help scientists understand complex brain diseases such as Parkinson’s and Alzheimer’s.
Scientists have designed a virus to send an enzyme to a precise location in the brain of a living mouse. The enzyme works by genetically tagging its neighboring proteins in a predetermined location.
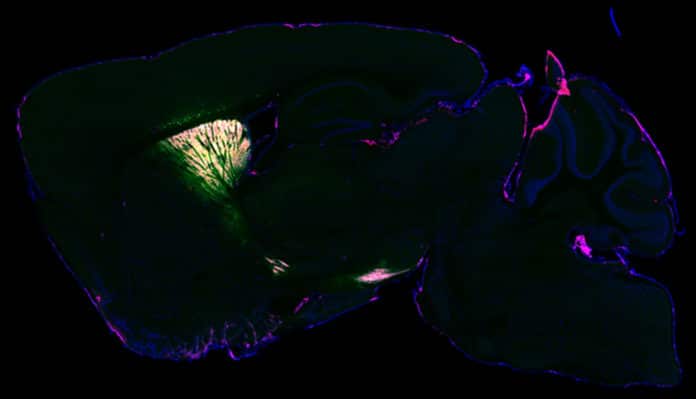
To validate their approach, scientists performed brain imaging with fluorescence and electron microscopy. They found that their technique took a snapshot of the entire set of proteins (or proteome) inside living neurons.
Northwestern’s Yevgenia Kozorovitskiy, a senior author of the study, said, “Similar work has been done before in cellular cultures. But cells in a dish do not work the same way they do in a brain. Also, they don’t have the same proteins in the same places doing the same things. It’s a lot more challenging to do this work in the complex tissue of a mouse brain. Now we can take that proteomics prowess and put it into more realistic neural circuits with excellent genetic traction.”
Vasin Dumrongprechachan, a Ph.D. candidate in Kozorovitskiy’s laboratory and the paper’s first author, said, “With our approach, we can start mapping the proteome of various brain circuits with high precision and specificity. We can even quantify them to see how many proteins are present in different parts of neurons and the brain.”
“Now that this new system has been validated and is ready to go. Hence, we can apply it to mouse models for disease to better understand neurological illnesses.”
“We are hoping to extend this approach to start identifying the biochemical modifications on neuronal proteins that occur during specific patterns of brain activity or with changes induced by neuroactive drugs to facilitate clinical advances.”
Journal Reference:
- Dumrongprechachan, V., Salisbury, R.B., Soto, G. et al. Cell-type and subcellular compartment-specific APEX2 proximity labeling reveals activity-dependent nuclear proteome dynamics in the striatum. Nat Commun 12, 4855 (2021). DOI: 10.1038/s41467-021-25144-y