Prostate cancer is the most common cancer and the second leading cause of death among men. The main challenge is to choose the best treatments for individuals at all stages of the disease.
To find out how aggressive somebody’s cancer is, specialists search for anomalies in slices of biopsied tissue on a slide. In any case, this 2D technology makes it hard to analyze marginal cases appropriately.
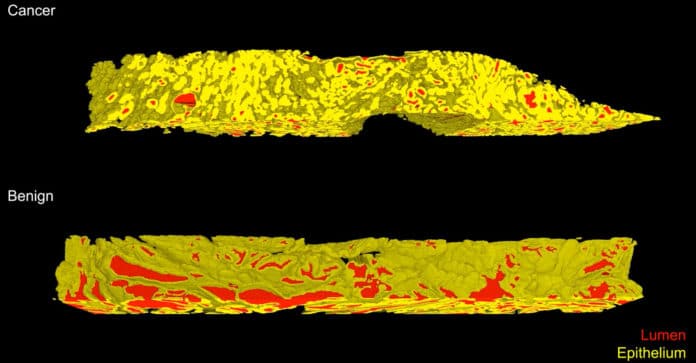
A team led by the University of Washington has developed a new, non-destructive 3D imaging method to improve risk assessment and treatment decisions. This method images entire 3D biopsies instead of just a slice. This analysis is based on interpretable glandular features and is facilitated by the development of image-translation-assisted segmentation in 3D (ITAS3D).
During the experiment, they imaged 300 3D biopsies taken from 50 patients — six biopsies per patient — and had a computer use 3D and 2D results to predict the likelihood of aggressive cancer. The 3D features made it easier for the computer to identify the more likely cases to recur within five years.
Senior author Jonathan Liu, a UW professor of mechanical engineering and bioengineering, said, “We show for the first time that compared to traditional pathology — where a small fraction of each biopsy is examined in 2D on microscope slides — the ability to examine 100% of a biopsy in 3D is more informative and accurate. This is exciting because it is the first of hopefully many clinical studies that will demonstrate the value of non-destructive 3D pathology for clinical decision-making, such as determining which patients require aggressive treatments or which subsets of patients would respond best to certain drugs.”
Scientists used the prostate samples from patients who underwent surgery more than ten years ago. They knew each patient’s outcome. They used the information to train a computer to predict the results. In the study, half of the samples contained a more aggressive cancer.
Scientists created 3D samples by extracting biopsy cores- cylindrically shaped plugs of tissue- from surgically removed prostates. They then stained biopsy cores to mimic the typical staining used in the 2D method.
The team then imaged each entire biopsy core. They used an open-top light-sheet microscope for this task, which uses a sheet of light to optically “slice” through and image a tissue sample without destroying it.
The 3D images provided more information than a 2D image. In particular, it offers detailed information on the complex tree-like structure of the glands throughout the tissue. These additional features increased the likelihood that the computer would correctly predict cancer’s aggressiveness.
Using AI methods, scientists managed and interpreted the large datasets this project generated.
Liu said, “Over the past decade or so, our lab has focused primarily on building optical imaging devices, including microscopes, for various clinical applications. However, we started to encounter the next big challenge toward clinical adoption: how to manage and interpret the massive datasets that we were acquiring from patient specimens. This paper represents the first study in our lab to develop a novel computational pipeline to analyze our feature-rich datasets. As we continue to refine our imaging technologies and computational analysis methods, and as we perform larger clinical studies, we hope we can help transform the field of pathology to benefit many types of patients.”
Journal Reference:
- Weisi Xie, Nicholas P Reder, Can F Koyuncu et al. Prostate cancer risk stratification via non-destructive 3D pathology with deep learning-assisted gland analysis. DOI: 10.1158/0008-5472.CAN-21-2843