The microscope is mainly used for imaging applications to analyze terabytes of data per day. These applications can profit from late advances in computer vision and profound learning. Now, in collaboration with robotic microscopy applications, Google engineers have assembled high-quality image datasets that separate signal from noise.
In “Assessing Microscope Image Focus Quality with Deep Learning”, researchers trained a deep neural network to rate the focus quality of microscopy images with higher accuracy than previous methods. They added the pre-trained TensorFlow model with plugins in Fiji (ImageJ) and CellProfiler, two leading open-source scientific image analysis tools to use with the graphical user interface or invoked via scripts.
Samuel Yang, Research Scientist, Google Accelerated Science Team, said, “Our publication and source code (TensorFlow, Fiji, CellProfiler) illustrate the basics of a machine learning project workflow: assembling a training dataset (we synthetically defocused 384 in-focus images of cells, avoiding the need for a hand-labeled dataset), training a model using data augmentation, evaluating generalization (in our case, on unseen cell types acquired by an additional microscope) and deploying the pre-trained model.”
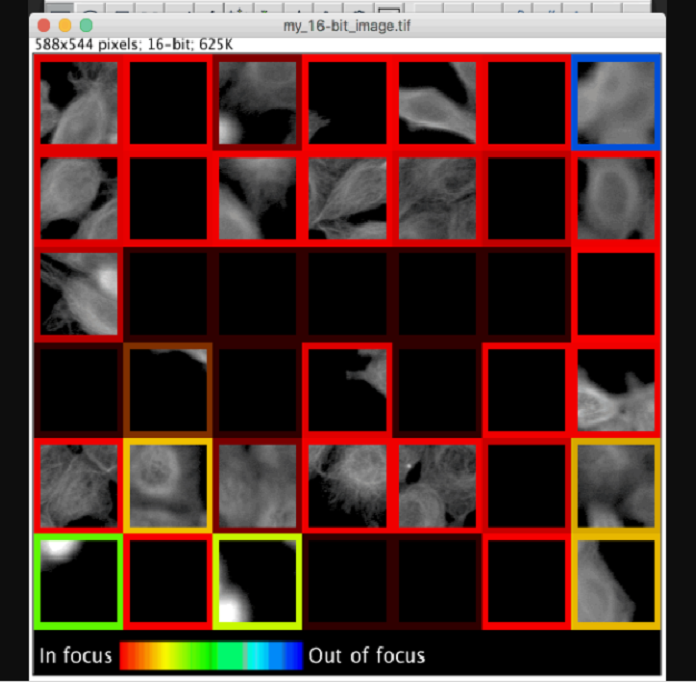
“Previous tools for identifying image focus quality often require a user to manually review images for each dataset to determine a threshold between in and out-of-focus images; our pre-trained model requires no user set parameters to use and can rate focus quality more accurately as well. To help improve interpretability, our model evaluates focus quality on 84×84 pixel patches which can be visualized with colored patch borders.”
“An interesting challenge we overcame was that there are often “blank” image patches with no objects, a scenario where no notion of focus quality exists. Instead of explicitly labeling these “blank” patches and teaching our model to recognize them as a separate category, we configured our model to predict a probability distribution across defocus levels, allowing it to learn to express uncertainty (dim borders in the figure) for these empty patches (e.g. predict equal probability in/out-of-focus).”
Journal Reference
- Yang, S.J., Berndl, M., Michael Ando, D. et al. Assessing microscope image focus quality with deep learning. BMC Bioinformatics 19, 77 (2018). DOI: 10.1186/s12859-018-2087-4