The liver is extensively time-regulated. This pattern is monitored by the circadian clock and biochemical signals, and eating rhythms.
The geography of the liver is interesting. It is divided into small repeating units called “lobules,” in which distinct zones perform different functions. This impressive spatial organization is called liver zonation.
Scientists have studied liver zonation statically by studying what each zone does independently of time, and vice-versa. And given how central the liver is in mammalian physiology, the two research approaches have to join efforts to understand how temporal and spatial liver programs interact.
In a new study, EPFL scientists, in collaboration with scientists from the Weizmann Institute of Science, monitored the spatial shifts of gene expression within liver lobules in relationship to the circadian clock.
Scientists analyzed the liver tissue in every individual cell. Interestingly, they studied almost 5000 genes in liver cells at several time points throughout the 24-hour day.
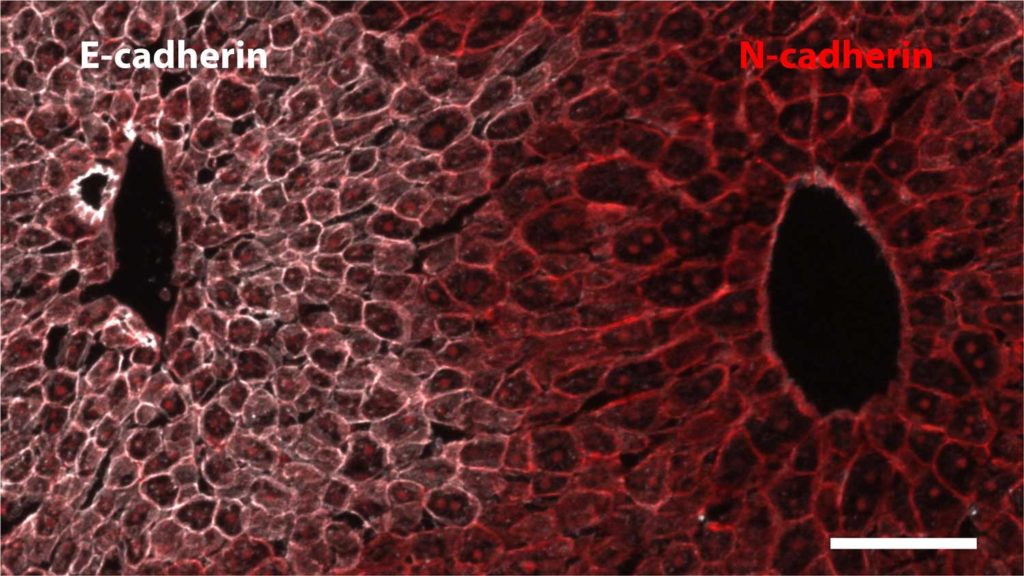
By statistically classifying the space-time patterns, scientists uncovered a model that can capture both spatial and temporal variations in messenger RNA levels (mRNA), a marker of gene expression.
The study revealed that many of the liver’s genes seem to be both zonated and rhythmic, meaning that they are regulated by their location in the liver and the time of the day. These dually regulated genes are mostly linked to key functions of the liver, e.g., the metabolism of lipids, carbohydrates, and amino acids, but also include a few genes that have never been associated with metabolism, e.g., genes related to chaperone proteins, which help other biomolecules change their 3D structure or even assemble and disassemble.
Professors Felix Naef at EPFL’s School of Life Sciences said, “The work reveals a richness of space-time gene expression dynamics of the liver, and shows how compartmentalization of liver function in both space and time is hallmark of metabolic activity in the mammalian liver.”
Journal Reference:
- Colas Droin, Jakob El Kholtei, Keren Bahar Halpern, Clémence Hurni, Milena Rozenberg, Sapir Muvkadi, Shalev Itzkovitz, Felix Naef. Space-time logic of liver gene expression at sub-lobular scale. Nature Metabolism 11 January 2021. DOI: 10.1038/s42255-020-00323-1