Scientists are taking a lot of interest in enzymes of microbes in order to find new ways to remove greenhouse gases from the atmosphere and turn them into useful carbon-containing compounds.
There are industrial procedures that do these reactions at high temperatures and high weights, and afterward, there’s this enzyme that can do a similar thing at room temperature.
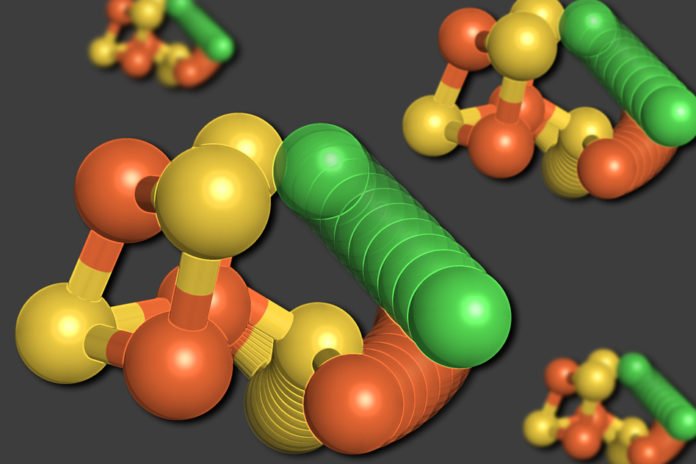
Now, MIT scientists have discovered a unique aspect of the structure of the “C-cluster” — the collection of metal and sulfur atoms that forms the heart of the enzyme carbon monoxide dehydrogenase (CODH). Instead of forming a rigid scaffold, as had been expected, the cluster can actually change its configuration.
Metal-containing clusters, for example, the C-cluster perform numerous other basic reactions in organisms, including part nitrogen gas, that is hard to recreate modernly.
Scientists started considering the structure of carbon monoxide dehydrogenase and the C-cluster around 20 years back, not long after she began her lab at MIT. They came up a structure for the compound utilizing X-ray crystallography, yet the structures weren’t exactly the same. The distinctions were in the long run settled and the structure of CODH was believed to be entrenched.
The task started with the hope of nailing down why the chemical is so sensitive to inactivation by oxygen and deciding how the C-clusters gets set up together.
The outcomes revealed two distinct structures for the C-cluster. The first was an arrangement they had expected to see — a cube consisting of four sulfur atoms, three iron atoms, and a nickel atom, with a fourth iron atom connected to the cube.
In the second structure, in any case, the nickel atom is expelled from the cube-like structure and replaces the fourth iron atom. The dislodged iron atom ties to a nearby amino acid, cysteine, which holds it in its new area. One of the sulfur atoms additionally moves out of the cube.
All of these movements appear to occur in unison, in a movement the researchers describe as a “molecular cartwheel.”
Catherine Drennan, an MIT professor of chemistry and biology said, “The sulfur, the iron, and the nickel all move to new locations. We were really shocked. We thought we understood this enzyme, but we found it is doing this unbelievably dramatic movement that we never anticipated. Then we came up with more evidence that this is actually something that’s relevant and important — it’s not just a fluke thing but part of the design of this cluster.”
According to scientists, this movement occurs upon oxygen exposure, helps to protect the cluster from completely and irreversibly falling apart in response to oxygen.
Douglas Rees, a professor of chemistry at Caltech, described the paper as “a beautiful study of a fascinating cluster conversion process.”
“These clusters have mineral-like features and it might be thought they would be ‘as stable as a rock. Instead, the clusters can be dynamic, which confers upon them properties that are critical to their function in a biological setting.”
Scientists are now searching for how cells assemble these clusters. Learning more about how these clusters work, how they are assembled, and how they are affected by oxygen could help scientists who are trying to copy their action for industrial use.
There is a great deal for concocting approaches to battle ozone-depleting substance amassing by, for instance, changing over carbon dioxide to carbon monoxide and after that to acetic acetate, which can be utilized as a building block for some sorts of helpful carbon-containing compounds.
Drennan said, “It’s more complicated than people thought. If we understand it, then we have a much better chance of really mimicking the biological system.”