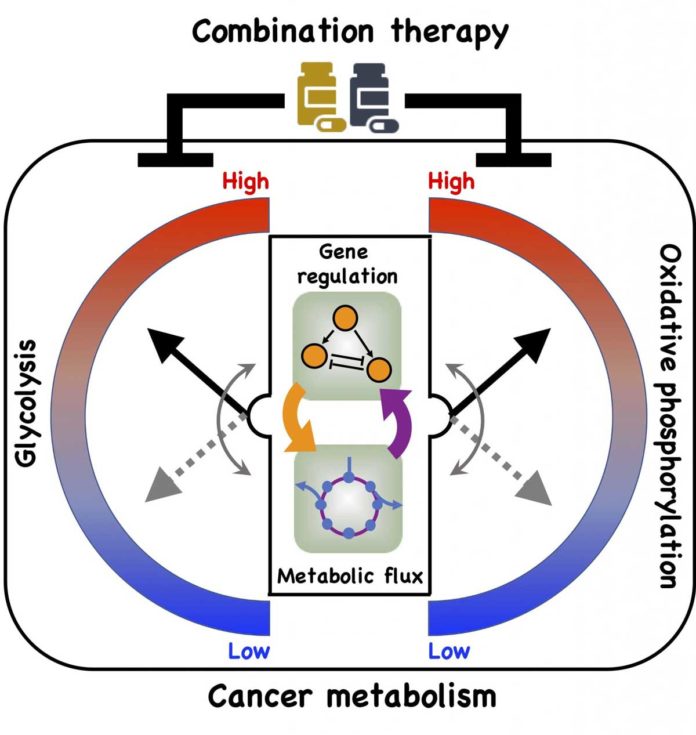
Metabolic plasticity enables cancer cells to switch their metabolism phenotypes between glycolysis and oxidative phosphorylation (OXPHOS) during tumorigenesis and metastasis. However, it is still largely unknown how cancer cells orchestrate gene regulation to balance their glycolysis and OXPHOS activities.
Rice University scientists are beginning to get a handle on how they survive hostile environments. Scientists have created a basic framework of how cancer cells — whether in tumors or as single cells — adapt when their attempts to metastasize are blocked by drugs or the body’s immune system. Understanding the cells’ strategies could someday help scientists design therapies that keep them in check.
Their model shows direct association between gene regulation and metabolic pathways and the way cancer cells take benefit of it to adapt to hostile environments, a process known as metabolic plasticity.
In particular, scientists observed oxidative phosphorylation (OXPHOS) and glycolysis, metabolic processes that provide cells with the energy and chemical building blocks they need to proliferate.
This for the first time, scientists have shown a direct connection between the activities of two protein players, AMP-activated protein kinase (AMPK) and hypoxia-inducible factor-1 (HIF-1), the master regulators of OXPHOS and glycolysis, respectively, with the activities of three major metabolic pathways: glucose oxidation, glycolysis and fatty acid oxidation.
José Onuchic said, “A lot of early cancer papers focus on the Warburg effect when cancer cells primarily use glycolysis even in the presence of oxygen. This is true, but it’s not like cancer cells give up on other mechanisms. The more aggressive they become, the more they are able to use any available choice to acquire energy. Our model shows how that’s possible.”
Postdoctoral fellow Dongya Jia said, “Only recently have people paid attention to OXPHOS. But they don’t really understand how cancer cells regulate these two metabolic phenotypes. We want to know how cancer cells orchestrate them. Since there is an extensive cross-talk between gene regulation and metabolic pathways, we think it’s necessary to simultaneously look at these two different aspects of cancer metabolism.”
Scientists noted, “Their model helped the team hone in on critical processes that traditional genome-scale metabolic models might miss. We start with simple models where we can figure out completely what’s going on, and then we add details to that scaffold without losing the basic understanding of how the system’s working.”
The model also subtleties associations that enable cancer cells to receive three stable metabolic states. One is a glycolytic state, described by high action of HIF-1 and high movement of the glycolytic pathway. The second is an OXPHOS state, described by high action of AMPK and high action of such OXPHOS pathways as glucose oxidation and unsaturated fat oxidation.
The third is a hybrid metabolic state characterized by high activity of AMPK and HIF-1 and of the glycolysis and OXPHOS pathways. The Rice model revealed the presence of both HIF-1 and AMPK can lead to the hybrid state that is difficult for current cancer therapies to address.
The researchers also found the hybrid metabolic state can be promoted by the stabilization of HIF-1 and the elevated production rate of mitochondrial reactive oxygen species (ROS) in cancer cells relative to normal cells. ROS are chemically active molecules that are important to signaling but at high levels can damage cells.
Kaipparettu’s Baylor team backed up the theory using gene expression data from breast cancer patients and metastatic triple negative breast cancer experimental models. Experimental evidence showed that repressing glycolytic activity in the cells activated AMPK and enhanced OXPHOS. The reverse was also true. But a combination of inhibitors that attacked both glycolysis and OXPHOS successfully eliminated the cells’ metabolic plasticity.
“We’re trying to push the field of metabolic modeling towards more flexibility, allowing for the decision-making processes we see in cells,” Levine said. “And here we’re coupling genes to metabolism in a way that’s rather novel.
“It’s still a limited view of all the metabolic pathways,” he said. “There are yet other possibilities that are not included in our model. We eventually need to tell a more complete story to really know what’s happening.”
Co-authors of the paper are former Rice postdoctoral researcher Mingyang Lu, an assistant professor at The Jackson Laboratory, Bar Harbor, Maine; postdoctoral associates Kwang Hwa Jung and Jun Hyoung Park of Baylor; and Rice alumnus Linglin Yu. Levine is an adjunct professor of bioengineering at Rice and a University Distinguished Professor at Northeastern University. Onuchic is the Harry C. and Olga K. Wiess Chair of Physics, a professor of physics and astronomy, of chemistry and of biochemistry and cell biology and co-director of the CTBP.
The new study appears in the Proceedings of the National Academy of Sciences.